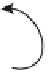Biology Reference
In-Depth Information
FIGURE 4.1
Transcription plays a
pivotal role in the regulation of gene
expression. (A) The central dogma in
molecular biology. (B) Different types of
networks regulate gene expression: TFs are
proteins that control gene expression by
interacting with the genome; non-coding
RNAs affect both the genome and the tran-
scriptome and RNA binding proteins affect
gene expression post-transcriptionally. (C)
Cartoon depicting transcriptional regulation
for an individual eukaryotic gene.
(A)
(B)
Genome
DNA
non-coding
RNAs
transcription
Transcription
factors
Transcriptome
RNA
translation
RNA binding
proteins
Proteome
Protein
(C)
general TFs
RNA
pol-II
regulatory TFs
exon
exon
TSS
cis
-regulatory module
basal promoter
eukaryotes have three different RNA polymerases: protein-
coding genes are transcribed by RNA polymerase II, while
RNA polymerase I and III transcribe rRNAs and tRNAs.
The most important mechanism of gene regulation is by
changing the levels of gene transcription. In addition to
RNA polymerase II, a set of general TFs is required for all
eukaryotic genes to be transcribed at a basal level
(
Figure 4.1
C). For each individual gene, this basal level of
transcription can be enhanced or reduced by regulatory TFs
(hereafter referred to simply as TFs) that are characterized
by the presence of a DNA-binding domain that directly
interacts with short DNA sequences located within gene
promoters, enhancers or other types of complex regulatory
sequences (see below). Through additional protein
domains, TFs interact with chromatin remodelers and/or
members of the basal transcriptional machinery to achieve
transcriptional activation or repression. Individual genes
are regulated by distinct sets of TFs, which leads to the
exquisitely specific spatiotemporal gene expression
patterns and levels that are required in biological processes
ranging from development to pathologies.
It is well known that TFs are of critical importance
during the development of complex multicellular organ-
isms. For instance, several TFs have been identified as
master regulators of the formation of particular organs and
tissues during development. Master regulators are classi-
cally defined as being necessary and sufficient to induce
organogenesis or other developmental processes. Exam-
ples include Pax6, a key regulator of eye development in
many different species
[2]
, and ELT-2, which is central to
intestinal development in the nematode Caenorhabditis
elegans
[3]
. TFs are important not only during develop-
ment, but also in adult organisms. Interestingly, most TFs
are expressed throughout the lifetime of an organism,
suggesting that many may play broad and important roles
in the responses
environmental or pathological cues. While some TFs, such
as master regulators, have been studied in detail using
biochemical and/or genetic approaches, the functions of
the vast majority of eukaryotic TFs remain unknown. This
is in part because most TFs likely have rather subtle
regulatory and biological effects compared to master
regulators, and may act redundantly with other TFs. As
a result, they are less likely to be identified in genetic
screens to function in a particular biological process.
Currently, many efforts aim to gain insight into how many
TFs interact together in GRNs to give rise to a desired
biological output.
GENE REGULATORY NETWORKS (GRN
S
)
GRNs are circuits composed of physical and/or regulatory
interactions between the two types of components that
orchestrate transcriptional outputs: TFs and cis-regulatory
elements (CREs) located in the genome. GRNs can be
represented as graphs that depict interactions between
genes (to which CREs are ascribed) and their regulators
(
Figure 4.2
). Network graphs are composed of nodes and
edges (see Chapter 9). GRNs are bipartite, which means
that there are two types of nodes: TFs and their target
genes. GRNs are also directional, as the edges indicate
a relationship from aTFto a target gene or CRE
(
Figure 4.2
). Different types of edges can be included in
GRNs. First, direct physical interactions between TFs and
DNA elements attributed to a gene (e.g., promoter, intron,
or more distally located cis-regulatory module or CRM)
can be included. The second type of edge that can be
included is a regulatory interaction (logical relationship),
which can be inferred from experiments that identify which
genes are positively or negatively affected upon loss (or
overexpression) of a TF. Before discussing GRNs in more
to stress and other physiological,