Biology Reference
In-Depth Information
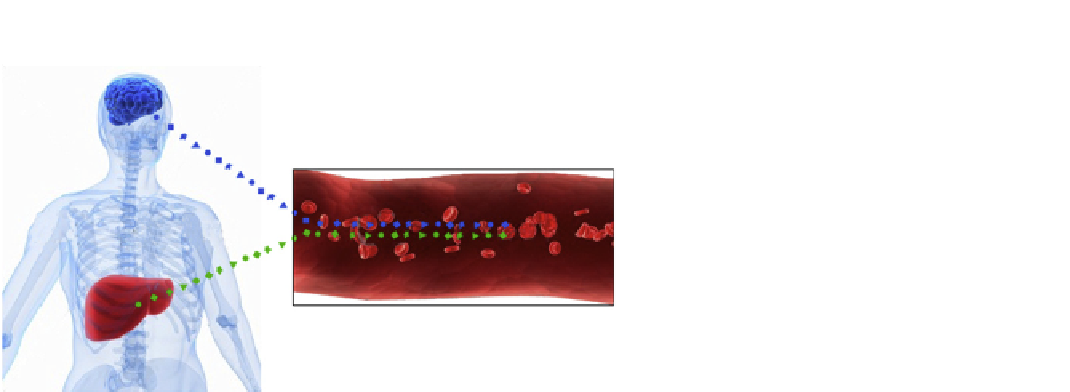
FIGURE 23.7
A diagram of organ-specific blood
fingerprints (collections of organ-specific proteins) from
the brain and the liver. For example, in a normal brain,
each of the proteins in the brain-specific blood fingerprint
will have one set of levels. In a diseased brain, the proteins
whose cognate networks have become disease perturbed
will change their levels. Since each disease leads to distinct
combinations of disease-perturbed networks an analysis of
the brain-specific protein fingerprints can distinguish
healthy from diseased brains, and if diseased can stratify
(e.g., distinguish from one another) the distinct types of
brain disease. Thus organ-specific brain fingerprints can
provide early detection, a stratification of different types of
disease and the ability to follow the progression of the
disease (not shown).
disease perturbed, and as the disease progresses more and
more networks are recruited to become disease perturbed.
(2) Two-thirds of the DEGs mapped into the four major
networks
these differentially expressed gene (DEG) transcripts
encoded proteins expressed in the blood, and we could
see the altered protein levels in the blood. This was an
example of presymptomatic diagnosis, a long-sought
keystone of early disease detection. However, these
DEGs were expressed in several different organs, so we
could not be certain of the location of the disease-per-
turbed process directly from observing protein
concentration changes in the blood. Second, in order to
obtain blood markers with organ-specific addresses, we
identified transcripts that were organ specific by deep
comparative
and the dynamics of these four networks
explained virtually every aspect of known prion disease.
(3) These four major networks were disease perturbed in
a serial manner: first, prion replication and accumulation;
second, glial activation; third, the degeneration of
neuronal axons and dendrites; and finally neuron
apoptosis. The importance of this observation is that if one
is interested in new approaches to diagnostics and thera-
peutics the initial disease-perturbed network is a logical
place to start. (4) The remaining one-third of the DEGs
identified six newnetworks that were heretofore unknown
to prion disease
e
transcriptome
analyses
across
40
þ
different organs in humans and mice (
Figure 23.7
).
From these analyses, through an examination of the
human and mouse blood protein databases, and exper-
imental mass spectrometry analyses, we were able to
identify about 100 brain-specific proteins in humans
and mouse. Of these, about 95% were orthologous
between the two species (the presumption is that they
will reflect similar activities in the two species), and
these proteins collectively constituted a brain-specific
blood fingerprint. We were able to show that some of
these brain-specific proteins could also be used for
presymptomatic diagnosis of prion disease in mice.
Additionally, brain-specific blood proteins encoded by
each of the four distinct networks exhibit concentration
changes in the blood in a serial manner consistent with
the order of disease perturbation of their cognate tran-
scriptional networks. These data demonstrate that we
will be able to assess both early disease detection and
disease progression from the blood.
In the organ-specific blood protein fingerprints each
individual protein assesses the behavior of its cognate
biological network, distinguishing normal functioning
from disease-perturbed functioning by changes in their
blood concentration levels. Because each disease per-
turbs different combinations of networks, the brain-
specific blood fingerprints will be able to distinguish
normal from disease and,
the so-called 'dark genes and networks
of prion disease' as identified by the global analyses of
normal and diseased brain transcriptomes. These insights
emphasize the importance of global analyses of the
transcriptomes. (5) These studies suggested new
approaches to blood diagnostics that are discussed below.
(6) For therapy it is obvious that the first and most prox-
imal prion-specific network should be re-engineered with
drugs to make it behave in a more normal manner and
hopefully abrogate the downstream consequences of this
pathological progression. It is clear that multiple drugs
will be required to re-engineer biological networks. We
are now learning how to re-engineer biological networks
with drugs inmicrobes, so that we can come to understand
the basic logic of the approach and then extend it to higher
model organisms (mice) before applying it to humans.
e
A holistic approach to disease will use blood as
a window for monitoring health (wellness) and
disease. A systems approach to blood diagnostics
emerged from two ideas arising from the prion studies.
First, some transcripts are expressed in their disease-
perturbed networks 8 weeks or more before the first
clinical signs (e.g., 10 weeks and 18 weeks, respec-
tively). We were able to demonstrate that several of
if diseased,
identify the