Biology Reference
In-Depth Information
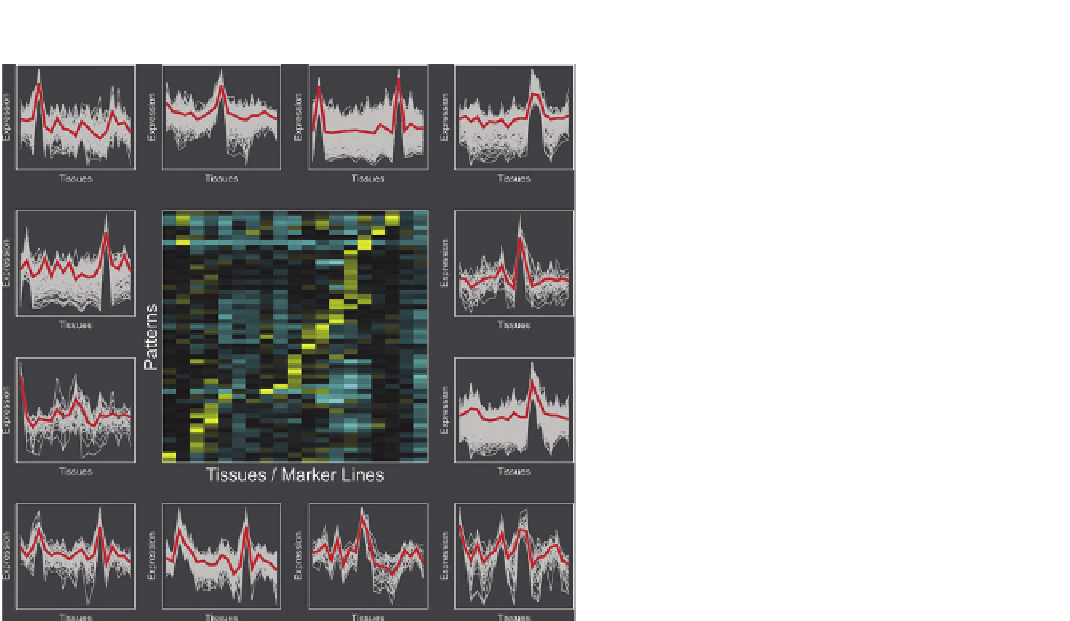
FIGURE 20.2
(continued).
(C)
root growth. The growth enhancement was due to an
increase in the number of cells undergoing division as well
as cells elongating to a larger size. When ectopically
expressed, this gene, named UPBEAT1 (UPB1), caused the
root to have a reduced growth rate with fewer dividing cells
and shorter cells in the elongation zone. To identify likely
targets of UPB1, which encodes a bHLH transcription
factor, microarray studies were performed on the mutant
and the ectopic expressor. This led to the identification of
a set of peroxidase genes as being repressed by UPB1
specifically in the elongation zone. Chromatin immuno-
precipitation followed by microarray analysis (ChIP-chip)
revealed that the promoters of these peroxidase genes were
bound directly by UPB1. Because peroxidases can control
reactive oxygen species (ROS), the ROS status in the root
was investigated using reagents that respond to different
ROS molecules. One type of ROS, hydrogen peroxide, was
found to be localized primarily in the elongation zone,
while another type, superoxide, was localized primarily to
the meristematic zone. The amount and localization of
these ROS changed dramatically in the upb1 mutant and
ectopic expressor. Manipulation of the different ROS with
small molecule agents provided evidence that they play
a central role in regulating the transition from cellular
proliferation to differentiation. Thus, a hypothesis based on
tissue-specific expression data led to the discovery of a key
transcription factor that acts through peroxidases to regu-
late the levels of reactive oxygen species, which in turn
control root growth rates
[10]
. Because UPB1 expression is
responsive to levels of hydrogen peroxide, there is a feed-
back loop that provides a means by which this subnetwork
could control ROS homeostasis.
A different type of hypothesis was generated from the
cell type-specific data set. The transcriptional regulation
that results in tissue-specific expression could be due to the
activity of transcription factors whose expression is
enriched in that tissue. Alternatively, tissue-specific
expression might be primarily controlled by transcription
factors whose expression overlaps in specific tissues, but
the expression of the factors themselves is not specifically
in that tissue. To determine which of these alternative
hypotheses was more likely to be correct, an experiment
was designed to identify the regulators for a set of genes
expressed in the vascular tissue and surrounding pericycle
(collectively known as the 'stele').
The yeast one-hybrid assay (see Chapter 4) was used on
full-length promoters of a set of 172 genes whose expression
was enriched in the stele
[11]
. These 172 promoters actually
belonged to the set of tissue-enriched transcription factors
that were tested for their ability to bind to these promoters. In
this way, both binding to other factor genes as well as binding
to their own promoters would be tested in the same assay. The
results were that approximately 25%of the genes were placed
into a network (
Figure 20.3
A), suggesting that a substantial
amount of tissue-specific expression results from regulation
by transcription factors enriched in that tissue. The surprise
came when the effects of mutating upstream factors on the
expression of downstream genes was determined. This was