Biology Reference
In-Depth Information
(A)
(B)
Two Alternative States
As suggested years ago by Nasmyth
[47,48]
, we can think
of the budding yeast cell cycle as an alternation between
two self-maintaining states: G1 is a state characterized by
unreplicated chromosomes and low activity of Clb-depen-
dent kinases; and S-G2-M is a state characterized by high
Clb-kinase activity and chromosomes in the process of
being replicated and aligned on the mitotic spindle. The
transition from G1 to S-G2-M involves commitment to
a new round of DNA replication and preparation for divi-
sion. The reverse transition involves partitioning the
replicated chromosomes to daughter nuclei (anaphase and
telophase) and cell division.
The G1 state is stabilized by inhibitors of Clb-kinase
activity, namely CKI and Cdh1. In the S-G2-M state, the G1
stabilizers are neutralized and the Clb-dependent kinases
are actively promoting DNA synthesis and mitosis
[7,17]
.
As illustrated in
Figure 14.5
A, there is mutual antagonism
between Clb-kinases and G1-stabilizers. The G1-stabilizers
neutralize Clb-kinase activity (CKI binds to and inactivates
Cdk1:Clb dimers, and Cdh1 promotes degradation of Clb
subunits). On the other hand, active Cdk1:Clb dimers
phosphorylate CKI and Cdh1, causing degradation of CKI
and inactivation of Cdh1. These mutually antagonistic
interactions create a basic bistable switch (see
Figure 14.6
and
Box 14.1
). In the 'neutral' position (SK
(C)
(D)
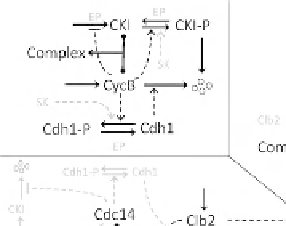
FIGURE 14.5
The molecular control systems of the budding yeast
cell cycle. (A) The fundamental bistable switch, created by the antago-
nistic interactions between the B-type cyclins
(CycB) and the
G1-stabilizers (CKI and Cdh1). SK
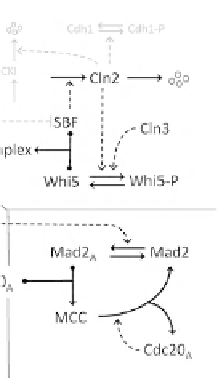
exit phos-
phatase. Four small circles are products of protein degradation. (B) The
Start transition. Production of Cln2:Cdk1, the starter kinase of panel A, is
controlled by the transcription factor, SBF, which is inactivated by
binding to a stoichiometric inhibitor, Whi5. Phosphorylation of Whi5
releases SBF. Notice the double-negative feedback loop between Cln2
and Whi5. Cln3 is a 'growth indicator' that triggers the Cln2 switch.
Notice also that Clb2-dependent kinase inactivates SBF, probably by
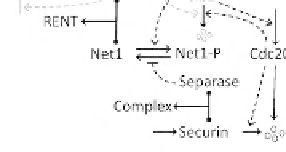
direct phosphorylation. (C) Exit from mitosis. Active Cdc20:APC
degrades securin and Clb2 (partially). Free separase degrades cohesin
rings (anaphase) and helps to release Cdc14 from inhibition by Net1.
Cdc14 is the exit phosphatase that activates Cdh1 and CKI. After Clb2
and other mitotic kinases are cleared by Cdh1:APC, Cdc20 is degraded,
Cdc14 is inactivated, and the cell reverts to the G1 steady state.
(D) Metaphase checkpoint. Clb2 and Cdc20 are components of both
networks C and D. The checkpoint protein, Mad2, is activated by
unaligned (tensionless) chromosomes, in a reaction that requires
Clb2-kinase activity. Active Mad2 binds to and sequesters Cdc20:APC in
the mitotic checkpoint complex (MCC). When all chromosomes have
come into alignment on the metaphase plate, the rate of activation of
Mad2 drops to zero, and the MCC starts to come apart in a reaction that is
accelerated by free (active) Cdc20:APC.
¼
starter kinase, EP
¼
¼
0in
Figure 14.6
), the CKI-Clb-Cdh1 dynamical network
can persist in either of two stable steady states: Clb inac-
tive, CKI and Cdh1 active (the G1 state), or Clb active, CKI
and Cdh1 inactive (the S-G2-M state). This picture is the
theoretical counterpart
[20]
of Nasmyth's intuitive notion
of 'alternative self-maintaining states'
[47]
.
In this theoretical framework the cell physiologist's G1/S
transition corresponds to a saddle-node bifurcation
(
Figure 14.6
, left), where the G1 branch of stable steady states
ends, and the control system switches irreversibly to the
S-G2-M branch of stable steady states. This transition is
promoted by a 'starter kinase' (
Figure 14.5
A), which helps
Clb-kinase to eliminate CKI and Cdh1. After the starter
kinase has flipped the switch, it is no longer needed to
maintain the system in the S-G2-Mstate. Similarly, theM/G1
transition ('exit from mitosis') corresponds to the reverse
saddle-node bifurcation (
Figure 14.6
, right), where S-G2-M
branch ends and the control systemswitches irreversibly back
to G1. An exit phosphatase opposes Clb-kinase activity and
helps the G1-stabilizers to reappear. Thereafter, it is no longer
needed to maintain the system in the G1 state.
The theoretical picture in
Figure 14.6
stands or falls on
the presumed bistability of the molecular interactions in
Figure 14.5
A. Bistability at the G1/S transition has been
confirmed experimentally by Cross
[49]
(see Figure 1 of
that paper). That irreversible exit from mitosis is due to
feedback loops rather than cyclin B degradation has been
¼
0 and EP
many cell cycle genes come in pairs: CLN1 and CLN2,
CLB1 and CLB2, CLB5 and CLB6
[46]
.Inourdiagrams
and models we lump these pairs together, i.e., 'Cln2'
represents both Cln1 and Cln2 protein pools, 'Clb2' both
Clb1andClb2,and'Clb5'bothClb5andClb6.Some-
times we lump together Clb1, Clb2, Clb5 and Clb6 as
'Clb'. Also, both Sic1 and Cdc6 function as stoichiometric
Cdk inhibitors in the mitotic cycle of budding yeast, so we
refer to both of them together as 'CKI'. CLN3 and BCK2
encode proteins that are jointly responsible for growth
sensitivity of the budding yeast cell cycle, and we usually
are referring to these proteins jointly as 'Cln3'
[7]
.The
expression of many cell cycle genes is controlled by
regulated transcription factors, but we shall refer explic-
itly only to the regulation of CLN1, CLN2, CLB5 and
CLB6 genes by SBF.