Biology Reference
In-Depth Information
TABLE 14.1
Glossary
d
cont'd
Two-parameter bifurcation
diagram
Plot of regions of distinct dynamical behaviors (e.g., monostability or bistability) in dependence on two
parameters of a dynamical system.
Ubiquitin
A small polypeptide (76 amino acids) that can be covalently linked to proteins in order to label the protein for
degradation by proteasomes.
Variable
A time-dependent component of a dynamical system, e.g., the activity of a cyclin-dependent kinase.
daughter cells are the chromosomes
e
the cell's genetic
material. In eukaryotic cells, the processes of chromosome
replication and partitioning are accomplished in separate
phases of the cell cycle (
Figure 14.1
): S phase (DNA
synthesis) and M phase (mitosis).
A eukaryotic chromosome consists of a linear, double-
stranded DNA molecule in close association with many
types of DNA-binding proteins. During S phase the DNA
molecule is carefully copied to produce two identical
double-stranded DNA molecules, which become similarly
decorated with DNA-binding proteins, to produce a pair of
'sister chromatids'. (Slight differences in the nucleotide
sequences of sister chromatids may arise from mistakes
e
mutations
e
in the copying process.) As sister chromatid
pairs are created during S phase they are physically bound
together by protein bands called 'cohesin rings'
[12]
. After
DNA synthesis is completed there is a gap (G2 phase)
during which the cell prepares for mitosis. G2 cells are
defined by having fully replicated chromosomes that have
not yet started the mitotic process.
The goal of mitosis is to separate the sister chromatids,
delivering one (and only one) copy of each chromosome to
each of the incipient daughter cells. Mitosis is a complex
and delicate process
[13]
. In prophase, the nuclear
membrane breaks down, the cytoskeleton rearranges to
form a bipolar spindle apparatus, and the replicated chro-
mosomes condense into highly compacted, X-shaped
bodies
[14]
. During prometaphase, microtubules emanating
from the poles of the mitotic spindle search for the
condensed chromosomes and bind to docking sites (kinet-
ochores) on the sister chromatids at the mid-zone of the X,
where the chromatids are still held together by 'centro-
meric' cohesins
[15]
. The goal is to attach the sister chro-
matids, by their kinetochore microtubules, to opposite
poles of the mitotic spindle. When properly attached, each
X is pulled to the midzone of the spindle (the metaphase
plate) where it resides, under tension
[16]
, until every
replicated chromosome is properly aligned on the spindle
(metaphase). At this point in time, the centromeric cohesin
rings are cleaved, allowing the kinetochore microtubules to
pull the sister chromatids to opposite poles of the cell
(anaphase). Nuclear membranes are then reassembled
around each of the segregated masses of chromosomes,
forming a binucleate cell (telophase), which then divides
down the middle to form two daughter cells, each with
a full complement of unreplicated chromosomes (G1
phase).
Progression through the mitotic cell cycle is charac-
terized by four crucial features
[17]
. First of all, to maintain
a constant number of chromosomes per cell from genera-
tion to generation, it is necessary that S phase and M phase
alternate,
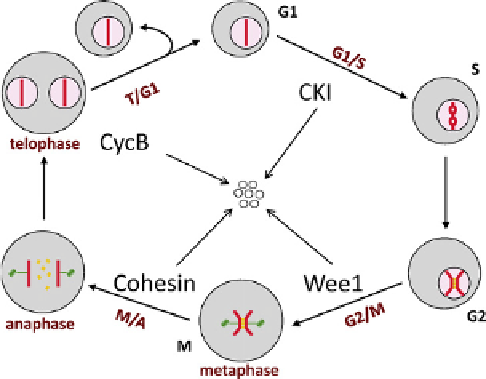
FIGURE 14.1
The eukaryotic cell cycle. Each 'cell' is bounded by
a membrane enclosing cytoplasm (gray) with a nuclear compartment
(pink) containing a representative chromosome (red bar). The chromo-
some goes through four distinct phases: G1, unreplicated; S, DNA
synthesis; G2, replicated; M, mitosis. During G2 phase the sister chro-
matids are bound together by cohesin rings (yellow). Mitosis consists of
distinct sub-phases: prometaphase (nuclear envelope breakdown, chro-
mosome condensation, spindle assembly), prophase (alignment of repli-
cated chromosomes on the spindle), metaphase (bi-orientation of all
chromosomes on the central plate), anaphase (cleavage of cohesin rings
and separation of sister chromatids to opposite poles of the spindle),
telophase (reassembly of envelopes around the daughter nuclei), cell
division. The four characteristic transitions of the cell cycle (G1/S, G2/M,
M/A and T/G1) represent four natural stopping points ('checkpoints')
where cell cycle progression can be halted if the cell detects any problems
with completion of essential functions of the pre-transition state. As the
cell passes each checkpoint it degrades a characteristic protein that had
been inhibiting the transition (CKI, Wee1, cohesin, cyclin B). The seven
small circles represent products of protein degradation.
i.e.,
that progress through the cell cycle be