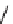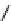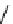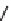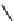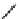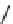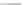Biology Reference
In-Depth Information
Fewer RNA than protein phenotypes have been char-
acterized experimentally, but computational analyses of the
relationship between RNA sequence and secondary struc-
ture point to much the same phenomenon. RNA genotypes
with the same secondary structure phenotype typically
form large genotype networks that extend far into, and
often all the way through, RNA genotype space
[87,109]
.
In sum, metabolic networks, regulatory circuits, and
macromolecules
enzymes with known sequence, tertiary structure, and
enzymatic function. It showed that small neighborhoods
around two proteins G1 and G2 that differ at fewer than
25% of their amino acids can contain sets of
of
new enzyme function phenotypes, such that the majority of
enzymatic functions found in
P1
and
P2
.
In sum, metabolic networks, regulatory circuits, and
macromolecules show two common qualitative properties
in the organization of their genotype space. Property 1 is
the existence of genotype networks that reach far through
genotype space. Property 2 is that small neighborhoods
around different genotypes typically contain different
phenotypes, even if the genotypes do not differ greatly.
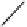
Figure 13.2
shows a schematic sketch of these properties.
The large rectangle in the figure stands for a hypothetical
genotype space. Each of the small circles stands for a single
genotype. The open circles correspond to genotypes that
share some hypothetical phenotype P (not shown).
Two genotypes are connected by a straight line if they are
P1
are not contained in
P2
show
a remarkable common property. Genotypes that have the
same phenotype are typically organized in large genotype
networks that reach far through genotype space. I will
return to genotype networks later, when I discuss their
significance for phenotypic variability.
very different kinds of systems
e
e
The Diversity of Neighborhoods
in Genotype Space
A second common property of the three system classes
emerges from the analysis of genotypic neighborhoods.
The neighborhood of a genotype is relevant for phenotypic
variability, because it contains genotypes that can be easily
reached from this genotype, that is, through one or few
small genotypic changes. For an analysis of phenotypic
variability, it is therefore useful to examine the spectrum of
phenotypes
that occur in a given neighborhood of
a genotype G1 that has some phenotype P. A simple
question is whether the spectrum of phenotypes in this
neighborhood depends on the genotype G1. More precisely,
consider two genotypes G1 and G2 with the same pheno-
type P and a given distance D. Denote as
P1
P1
and
P2
the sets
of phenotypes (different
from P)
in their
respective
neighborhoods. How different is the set
P1
from the set
P2
?
That is, are most phenotypes in
?Or
are most of these phenotypes unique to the neighborhood of
G1, in the sense that they do not also occur in the neigh-
borhood of G2?
In metabolism, one finds that the neighborhoods of two
metabolic genotypes G1 and G2 sampled at random from
the same genotype network contain mostly different novel
phenotypes. In other words, the set
P1
also contained in
P2
of new phenotypes
in the neighborhood of G1 is very different from the set
P1
P2
of new phenotypes in the neighborhood of G2. This holds
regardless of the specific genotypes G1 and G2, as well
as regardless of the specific phenotype P that they have
[54,57]
. The situation in regulatory circuits is not much
different. There, small neighborhoods around two circuits
G1 and G2 may contain sets of phenotypes
FIGURE 13.2
A highly simplified schematic of the structure of
a genotype network and the new phenotypes near it. See text for
details. Note that genotype networks are objects in a high-dimensional
genotype space with counterintuitive geometric properties. Also, actual
genotype networks contain an astronomical number of members. Indi-
vidual genotypes may have hundreds to thousands of neighbors, only few
of which can be shown. In addition, each of the genotypes shown in
different colors is also part of a vast genotype network that is not shown. A
figure like this can thus merely provide a modicum of intuition about the
organization of genotype space. (Adapted from
[10]
. Used with permission
from Oxford University Press.)
that
differ in the majority of their phenotypes, even for circuits
whose genotypes differ little, that is, in no more than 20%
of their regulatory interactions
[83]
. Much the same holds
for protein and RNA molecules
[87,92,110,111]
.For
example, a recent analysis studied more than 16 000
P1
and
P2