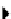Biology Reference
In-Depth Information
(A)
(B)
TRANSCRIPTION FACTORS
glc
TF1
TF2
TF3
atp
adp
g6p
f6p
atp
G1
G2
G3
G4
G5
adp
GENES
fdp
(C)
()
dhap
g3p
nad+
nadh,h+
13dpg
adp
atp
3pg
2pg
h2o
pep
adp
atp
pyr
(D)
glc
atp
adp
hexokinase
g6p
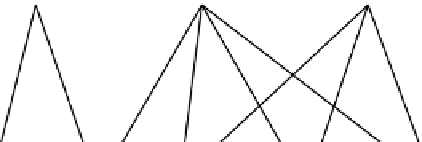
FIGURE 12.6
The components of the OME-matrix. (A) In a transcriptional regulatory network (TRN), transcription factors (TF) can modulate the
expression of various genes. Activation is indicated by arrows, and repression is shown with horizontal bars. (B) Genes that are modulated by TFs often have
gene products with important functions that depend on each other, such as with enzymes of the glycolytic metabolic pathway. (C) Transcription and
translation link transcription regulation and enzyme function. An overview of transcriptional and translational machinery systems demonstrates the
complexity of this link. Illustration used with permission from
[153]
. (D) The components each of these processes can be described mathematically, forming
an OME-matrix including transcription regulation ('O' for operon), metabolism ('M'), and transcription/translation ('E' for expression). This will allow for
the computation of how the processes work together as shown for a single GPR. The transcription factor binding to the ORF induces transcription of
hexokinase mRNA, which is translated by E-matrix components. The hexokinase enzyme then catalyzes the conversion of glucose to glucose-6-phosphate.
O-Matrix Reconstructions
The process behind reconstructing the transcriptional
regulatory network (TRN) is quite different from that of
metabolism or transcription and translation, owing to the
nature of the network. Where the latter networks can be
described via a series of stoichiometric equations for
component reactions, TRNs consist of a series of interac-
tions (either activation or repression) between transcription
factors (TFs) and their binding sites
[3]
. A common mode
of determining connectivity between TFs and their binding