Biology Reference
In-Depth Information
(A)
Metabolic engineering
6
6
0
0
2
2
2
6
ORF1 ORF2
?
ORF4
Predicted: No Growth
Experimental: Growth
ATCGTTCG
GCGCTAAT
GTATAGGC
ORF3
ORF1 ORF2 ORF3 ORF4
Predicted: Growth
Experimental: Growth
GENOME SCALE
RECONSTRUCTION
: Essential
: Conditionally Essential
: Non-essential
MODELS
Liver
Skin
Neuron
Spleen
/ / : Coupled Reactions
(D)
Network analysis
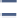
FIGURE 12.5
Applications of genome-scale metabolic reconstructions. (A) Computational algorithms can be used to determine combinatorial knockout effects on the production of a desired
small-molecule product. Furthermore, some methods can make predictions of genetic and environmental perturbations that will help improve product yield. Blue numbers correspond to flux through
a reaction, and the effect of a double KO to force flux to produce a desired metabolite is shown. (B) Biological discoveries are made as incorrect predictions are reconciled with experimental data. For
example, missing reactions can be predicted, thus guiding detailed biochemical studies to identify genes for orphan and missing reactions. (C) Phenotypic behavior is used both as a predictive tool (e.g.,
gene essentiality or adaptive laboratory evolution) and as a method to guide model improvements. (D) Reconstructions can be used for network analysis, thereby generating hypotheses about network
behavior (e.g., to predict coupled reactions or co-regulated genes). (E) High-throughput data can be used as a source for additional constraints or aid in the construction of tissue specific models, thereby
adding value to omic data. (F) Multi-cell and community relationships can be modeled by integrating multiple metabolic models allowing for simulation of phenomena such as symbiosis, competition, or
pathogenicity.