Biology Reference
In-Depth Information
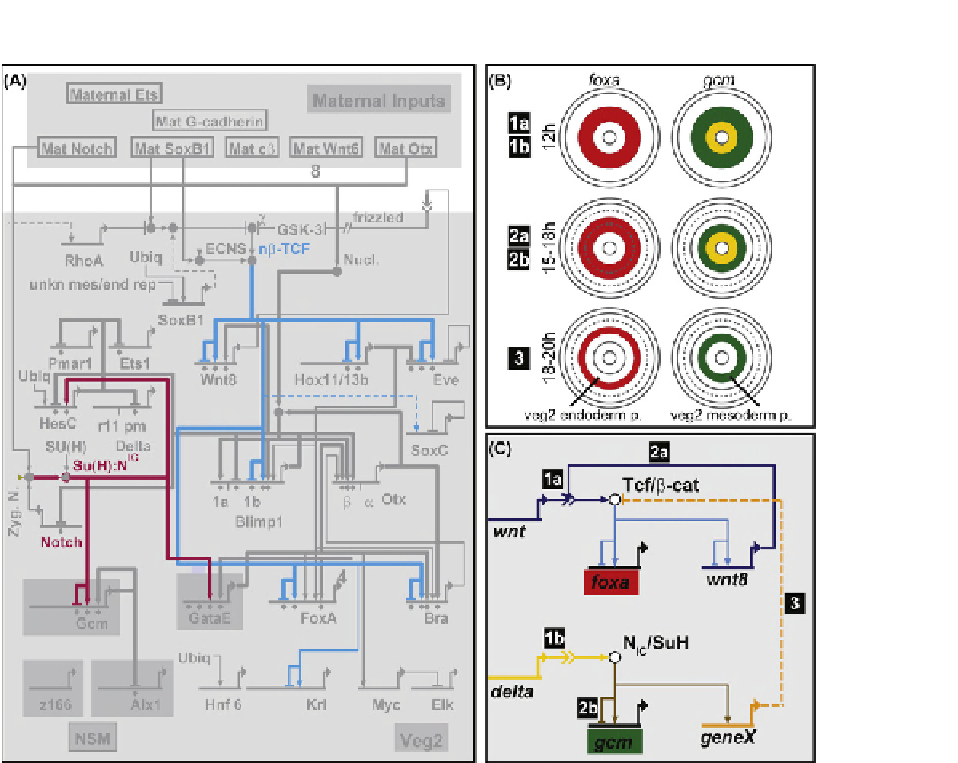
FIGURE 11.5
Endoderm/mesoderm cell fate specification in the sea urchin embryo. (A) Initiation of two non-interacting networks of regulatory
gene expression within the same precursor cells early in the process of specification of endoderm and mesoderm
[53]
. A mesodermal GRN is initiated in
response to Notch signaling (red GRN linkages); the endodermal GRN is initiated in response to Wnt signaling (blue GRN linkages). Cis-regulatory
genomic hardwiring determines which regulatory gene responds to which signal transduction system. (B) Diagrammatic portrayal of progressive spatial
separation of endodermal and mesodermal regulatory states. Red indicates expression of the endodermal GRN, here represented by the foxA regulatory
gene. Green indicates expression of the mesodermal GRN here represented by the gcm regulatory gene. Yellow indicates the location of the skeletogenic
cells and their expression of the Delta signaling ligand. At 12h a single ring of cells is precursor to both mesodermal and endodermal fate. At 15
e
18 h
a radial cleavage separates this domain into two rings of cells, of which only the inner ring now expresses the mesodermal GRN. At 18
e
20h, mesodermal
and endodermal fates are spatially separated with the terminal extinction of the endodermal GRN in the inner ring. (C) Network subcircuit responsible for
activation of mesodermal and endodermal GRNs, here represented only by the foxa and gcm genes. The Notch dependent linkage which extinguishes Tcf
function and hence endoderm gene expression is indicated.
specification for an episode of embryogenesis, or the
formation of a body part, we will have taken a giant step
toward explaining that aspect of development. We have
comprehensive explanations couched in GRNs for pair rule
and gap gene expression patterns
[39,73]
, and for meso-
derm diversification
[74]
in Drosophila; and for much of
pregastrular specification in the sea urchin embryo
[14,15,21,53]
. Diverse specification GRNs have become
available for embryonic processes in other model systems
as well as for the development of at least some adult
vertebrate and arthropod body parts (for reviews, see refs.
[19,22,24]
. As proof of principle, the best-known portions
of the sea urchin GRNs summarized in
Figure 11.2
generate
explanations for specification that do indeed fulfill all
the criteria for explanation listed above. Finally,
explanations we have for these specification functions
provide a pathway causally linking the regulatory genome
to regulatory states in embryonic space. Therefore we can
understand both developmental specification in terms of the
genomic program, and the regulatory genomic code in
terms of its functional output, regional embryonic specifi-
cation. There can be no satisfying explanation of devel-
opment that is not rooted in the genomic information that
encodes its control program, and in that sense specification
GRNs provide the root for all downstream explanatory
mechanisms.
But development scarcely ends with specification and the
establishment of regulatory state patterns. By the same
criteria, we lack any example of system-wide comprehensive
explanations of developmental processes that carry all the
the