Biology Reference
In-Depth Information
as PTM site occupancy together with kinetics upon
perturbation.
Even after correctly and comprehensively measuring
the phosphorylation changes upon cellular perturbation, the
question remains which kinase or kinases are responsible
for a given phosphorylation site. A variety of combinations
of quantitative phosphoproteomics and chemical genetics
approaches can answer this question by identifying direct
kinase substrates. For instance, this can involve controlled
inhibition of a genetically engineered cellular kinase by
a small molecule
[167]
. In an alternative approach, phos-
phorylation patterns in 124 kinase and phosphatase yeast
deletion strains have been measured to globally extract
kinase
different peptides, specialized MS strategies may have to be
used, such as top-down proteomics
[172,173]
.
As described above, MS-based PTM analysis is
uncovering an unexpectedly large extent and diversity of
PTMs that occur on multiple but specific residues on most
proteins. These large-scale PTM studies now serve as an
information-rich resource to the community. For example,
biological researchers can focus on regulatory PTM sites in
high-quality MS data for their proteins or processes of
interest. The data can also be used to investigate basic
characteristics of particular PTMs, such as their evolu-
tionary conservation
[154,174,175]
and preferential local-
ization across secondary structures of proteins
[154,176]
.
In addition, in vivo maps of many PTMs are beginning
to emerge
[150,157,177,178]
and a first example of large-
scale PTM quantification in a mouse organ after perturba-
tion has been described
[179]
. This is now unlocking the
opportunity to study PTM dynamics in tissues to charac-
terize the physiological or pathological responses of
different organs in mammals. With their key roles in
cellular control, MS-enabled PTM signatures also hold
great potential as prognostic and therapeutic biomarkers.
substrate relationships
[168]
. To understand the
circuitry that underpins cellular information flow, the
changes in PTM dynamics can additionally be overlaid
with direct protein
e
protein interaction datasets such as
those of kinases and phosphatases from yeast
[169]
.
Clearly, it would also be important to understand how the
dynamic kinase
e
substrate interactions vary under different
growth and stress conditions.
From a cellular control perspective, a significant
increase in information content can be achieved if many
proteins are multiply modified, especially if these PTMs
acted combinatorially. In fact, it has become increasingly
clear that a number of cellular processes are regulated by
PTM cross-talk, as exemplified by phosphorylation and
ubiquitylation
[170]
. Another example is the intimate
interplay of different histone modification marks in the
histone code, which represents one of the most important
epigenetic regulatory mechanisms governing the structure
and function of the genome
[171]
. To understand such PTM
cross-talk codes that mediate cellular control, MS-based
proteomics is an excellent large-scale method. However,
owing to the fact that correlating PTMs may occur on
e
OUTLOOK AND FUTURE CHALLENGES
As detailed in this chapter, MS-based proteomics is
a technology-driven discipline that has made tremendous
progress during recent years. These advances affect the
entire proteomics workflow, starting with sample prepara-
tion and ending with computational proteomics. The advent
of high-resolution high-accuracy MS data, combined with
sophisticated quantification strategies, has been especially
important in obtaining biologically relevant information
from MS-based proteomics. This technology has now
clearly become the method of choice for
studying
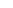
A: Genomics
B: Transcriptomics
C: Proteomics
protein
interaction
gene regulation
post-
ncRNAs
translational modification
replication
transcription
transcription
protein
localization
proteins
signal
transduction
splicing
epigenetics
genes
translation
gene
products
protein turnover
many genomes
in parallel
transcriptomes
in time
proteomes in time and space
protein interaction
post-translational modification
protein localization, dynamics and turnover
#1
#2
#3
#4
FIGURE 1.6
Unique contribution of different 'omics' technologies to systems biology. A: Genomics investigates the sequences of genomes and
their epigenetic marks for many genomes in parallel, but does not provide direct information about the fate of the gene products. B: Transcriptomics
measures the gene expression program and allows the comparison of changes of gene expression in different cellular states or over time. C: Proteomics
strives to provide a complete picture of all proteins, the primary agents of cellular processes. Proteins can be monitored over time and with sub-cellular
resolution along with their post-translational modifications, interactions and turnover.