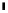Biology Reference
In-Depth Information
BRX1, DIS3,
ERB1, MTR3,
RLP7, RRP42,
RRP45, RRP46,
SKI6, YPR142C
rDNA clusters
Cytoplasm
Transcription
NCL1
NPM1
Nucleolus
47S pre-rRNA
5.8S rRNA maturation
(
P
= 3.0
×
×
10
−
7
)
Nucleus
Processing
18S
28S
5.8S
NOC2, NOP2,
NOP15, RPL24
5S RNA
MEX67, MTR2,
NIC96, NUP49,
NUP57, NUP85,
NUP145
Assembly
LSU precursor
(
P
= 1.5
××
10
−
3
)
Pre-60S
RPL
RPS
Pre-40S
nuclear pore (
P
= 3.4
××
10
−
10
)
subunit export from nucleus (
P
= 6.4
××
10
−
6
)
Export
Export
Polypeptide
tRNA
GTP
80S ribosome
Pre-40S
AAAAA
Pre-43S
elF2
5'm7GTP-cap
mRNA
elF3
elF4E
Protein synthesis
GCD11, NIP1,
SUI2, SUI3,
TIF35, YPL238C
RPL5, RPL10, RPL17A, RPL18A,
RPL30, RPL33A, YBR190W,
YLR076C, YPL142C
cytosolic LSU (
P
= 7.4
××
10
−
7
)
multi-EIF complex (
P
= 2.1
××
10
−
5
)
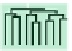
FIGURE 8.3
Co-fitness reveals the road to translation: translational regulation by genes involved in ribosomal biogenesis. Genes were clustered
hierarchically based on co-fitness, i.e., similarity between their corresponding deletion strain signatures, where each signature contains HIPHOP
measurements of the strain under ~3500 chemical perturbations conditions. Here we show a branch of the gene dendrogram that includes several clusters,
highlighted in colored boxes, which are associated with ribosome biogenesis. For each highlighted cluster, select gene ontology biological processes and/
or protein complexes that are significantly enriched amongst the member genes are shown in a box of the same color as the cluster box (corrected
p
0.01), next to a schematic of ribosome biogenesis in the cell. The multiple-test corrected p values measuring the significances of the enrichments are
shown in parentheses. Further, the genes in the highlighted clusters are listed above the corresponding enrichment boxes. The biogenesis of the ribosome
machinery is a highly coordinated process, which is composed of the synthesis and import of ribosomal proteins into the nucleus, synthesis and processing
of ribosomal RNA (rRNA), assembly of ribosomal proteins and subsequent transport of the mature subunits into the cytoplasm. Most of these events take
place in the nucleolus, except for 5S rRNA synthesis (which occurs in the nucleoplasm) and synthesis of ribosomal proteins (which occurs in the
cytoplasm). The basic translation machinery is composed of ribosomal subunits, messenger RNAs (mRNAs), transfer RNAs (tRNAs), and translational
initiation and elongation factors. First, the initiation factors eIF2, eIF3, tRNA and GTP are incorporated into a 40S ribosomal subunit to form a 43S
complex. Second, eIF4e is recruited into the 43S complex to form a 48S complex with mRNA. Finally, a 60S ribosomal subunit and the 48S subunit form
the final 80S complex. Abbreviations: eIF, eukaryotic initiation factor; LSU, large subunit of ribosome; rRNA, ribosomal RNA. [Adapted by permission
from Macmillan Publishers Ltd: Nature Reviews Cancer
[72]
copyright (2010)]
encoded by MDR genes include cytochrome P450s that
metabolize (~75% of all) drugs and to a much lesser extent
multi-drug transporters, many belonging to the protein
superfamily ATP-binding cassette transporters (ABC-
transporter) that catalyze the active extrusion of a large
number of unrelated small molecules out of the cell. From
the few existing datasets that are large enough, yeast che-
mogenomic profiles identified a set of genes that, when
deleted, are sensitive to multiple chemical perturbations
[21,26,47]
. Within this context, MDR genes have been
generally defined by the corresponding deletion strains
exhibiting a significant fitness defect to ~10
20% of the
small molecules tested. Given the fact that yeast is notorious
for its pleiotropic drug resistance (PDR) transporters and
general ability to resist chemical stress, though many are
redundant, we expected to identify PDR transporters as
MDR genes by our criteria; instead, we identified surpris-
ingly few, consistent with the findings of others
[21,26,47]
.
e