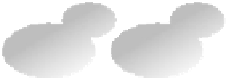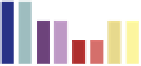Biology Reference
In-Depth Information
1. Pool tagged deletion strains
4. PCR-amplify uptags and downtags
Uptag PCR
Downtag PCR
2. Grow deletion pool in condition of choice
5. Hybridize PCR products to barcode microarray
6. Analyze data
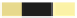
Starting sample
Growth sample
3. Purify genomic DNA
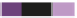
Tag
Tag
Analyzed data
Deletion strain
FIGURE 8.1
Description of the competitive growth assay. Fitness profiling of pooled deletion strains involves six main steps.
1.
Strains are first
pooled at approximately equal abundance.
2.
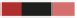
The pool is grown competitively in the condition of choice. If a gene is required for growth under this
condition, the strain carrying this deletion will grow more slowly and become underrepresented in the culture (red strain) over time. Similarly, resistant
strains will grow faster and become overrepresented (blue strain).
3.
Genomic DNA is isolated from cells harvested at the end of pooled growth.
4.
Barcodes
are amplified from the genomic DNAwith universal primers in two PCR reactions, one for the uptags and one for the downtags.
5.
Resulting PCR products
are hybridized to a barcode microarray that quantitates the tag sequences, and therefore the relative abundance of each strain..
6.
Tag intensities for the
treatment sample are compared to tag intensities for a control sample to determine the relative fitness of each strain. (Figure originally published in
[12]
).
The complete yeast deletion collection, often referred to
as the Yeast KnockOut (YKO) collection, is comprised of
four sets of strains: ~5000 haploids of mating type a, ~5000
haploids of mating type
construct and confirm each deletion strain. The construc-
tion strategy to make the knockout cassette modules used
an elegant design devised by Davis (the proof-of-concept
first demonstrated in bacteria
[13]
) that included the
incorporation of two molecular tags or 'barcodes' (the
second acting as a fail-safe) into each deletion strain. These
molecular barcodes, comprising 20 bp DNA sequences that
serve as unique strain identifiers, not only ensured against
strain mix-up, but allowed the function of each gene to be
interrogated in parallel by mixing them together. Specifi-
cally, in each competitive growth or 'fitness profiling' assay
(
Figure 8.1
) a single culture is first inoculated with an
approximately equal number of cells from each deletion
strain. The culture is then grown in a condition of interest
and samples are collected at several
a
, ~5000 homozygous deletion
strains and ~6000 heterozygous strains. The additional
~1000 heterozygous strains represent the ~18% of genes
that were determined to be essential (to unperturbed cells).
To construct the yeast deletion collection, tens of thousands
of oligonucleotides were necessary and the associated cost
would have been prohibitive had they been purchased from
commercial sources. Owing to the development of the first
homemade 96-well oligonucleotide synthesizers at the
Stanford Genome Technology Center, however, the oligo-
nucleotides could be made at dramatically lower cost.
Whereas most of the scientific community have realized the
significance of the YDP, few appreciate that the project was
contingent on the 96-well oligonucleotide synthesizer,
a technology that had already been dismissed by the
National Institute of Health (NIH) and by industry as
a technology in search of an application. The 96-well
synthesizer produced the eight unique primers required to
time points over
a period of 5
20 generations. At the end of the experiment,
genomic DNA is extracted from each sample and the
molecular barcode tags are amplified in two PCR reactions
(
Figure 8.1
). The abundance of each deletion strain is
quantified by hybridizing the resulting PCR products to an
oligonucleotide array carrying the complementary barcode
e