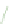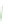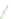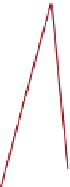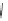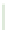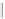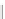Biology Reference
In-Depth Information
(A)
“Between-pathway”
“Within-pathway”
A
X
A
X
A
X
A
X
D
D
D
D
Y
Y
Y
Y
E
E
E
E
B
B
B
B
C
Z
C
Z
C
Z
C
Z
F
F
F
F
function
function
function
function
function
function
function
function
(B)
Structures
within modules
Structures
within modules
Structures
between modules
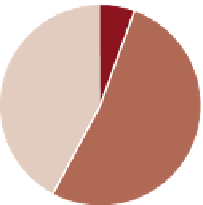
6%
Unstructured
3%
Unstructured
Structures
between modules
16%
42%
52%
81%
Negative
genetic interactions
Positive
generic interactions
protein
complex
1
protein
complex
2
protein
complex
3
protein
complex
4
(C)
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
X
E
protein
complex
1
A
A
D
B
C
C
F
D
E
H
protein
complex
2
B
F
G
X
X
G
H
I
I
J
protein
complex
3
M
J
K
L
N
M
L
K
protein
complex
4
O
N
O
Protein-protein interaction
Positive genetic interaction
Negative genetic interaction
Non-essential gene
Essential gene
Uncharacterized gene
Negative genetic
interaction
Positive genetic
interaction
No interaction
Positive genetic interaction
Negative genetic interaction
Essential gene
Uncharacterized gene
Structured
Structured
Unstructured
Unstructured
X
X
FIGURE 6.3
Genetic interaction network structure: within- and between-pathway structures. (A) Negative interactions (red lines) can arise from
the disruption of parallel pathways converging on a common process ('between-pathway' genetic interactions), or can occur between pairs of genes
within the same essential pathway or functional module ('within-pathway' genetic interactions). Semicircles depict conditional or hypomorphic
(partially functional) alleles of essential genes. Thick black lines indicate wild-type function, while thin black lines indicate reduced function. Grey
arrows indicate loss of function. (B) The fraction of negative and positive genetic interactions spanning within- and between-pathway structures, as well
as unstructured genetic interactions. (C) Properties of genetic interactions. Schematic representation illustrating the complexity of within- and
between-pathway structures. Both positive (green) and negative (red) interactions can exist as unstructured interactions, connect between two functional
modules, or connect members within the same functional module. Circular and diamond shaped nodes represent non-essential and essential genes,
respectively. The highly organized modular nature of the yeast genetic interaction network helps identify distinct groups of functionally related genes
and how these different modules relate to one another. Although the vast majority (~98%) of genetic interactions (both negative and positive) tend to
occur between pathways and protein complexes, negative interactions can connect members of the same essential protein complex while positive
interactions can occur between members of non-essential complexes. Dashed lines indicate physical interactions. Similar to the network representation,
two-dimensional clustering reveals distinct 'blocks' of genetic interactions that define functional modules. Interaction blocks highlighted along the
diagonal axis of the matrix correspond to within-pathway structures while off-diagonal blocks represent between-pathway modules. Modified from
[120]
.