Biology Reference
In-Depth Information
RBS
1
D
MIN
2
1
Protein
RBS
2
3
Fig. 2.5
The success definition of metaPocket. Ligands are illustrated as dotted lines and marked
by 1, 2 and 3. First all the ligands bound on protein surface are clustered by their spatial distance
using cutoff value DMIN. Here two real ligand binding sites are shown in circle and marked
as RBS1 and RBS2 with three ligands. In RBS2, the solid spheres in different size are shown.
The smaller spheres indicate the pocket sites from single methods and the bigger sphere indicates
the meta-pocket site of metaPocket. If these sites are within 4 Å to any atom of the ligand, then this
real ligand-binding site is successfully detected
cluster, are the potential ligand binding residues. The surface residues are defined
using the NACCESS program whose relative solvent accessible surface area is more
than 20%.
2.4
Evaluations of LIGSITE
csc
, MetaPocket
and Other Approaches
To evaluate and compare metaPocket with other single methods fairly, the same
performance measurement and data-set should be used. It is noted that for some
proteins in the data-sets we used here, more than one ligand is bound. These ligands
might be separated in different pocket sites but sometimes occupy the same region
on protein surface, for example, those co-factors and substrates. As illustrated in
Fig.
2.5
, first, we define the real ligand binding sites (RBS), which are those regions
on protein surface where one or more ligands are bound. If two ligands are closed









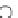

Search WWH ::

Custom Search