Biology Reference
In-Depth Information
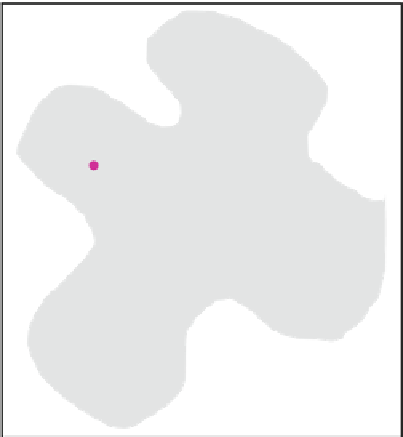
Protein atom
Grid point
Pocket site
Scan direction
Fig. 2.1
The detailed algorithm of LIGSITE
csc
. The protein is projected into a 3D grid (here 2D)
and the grid points located at the pocket sites are identified by scanning seven directions (four
directions for 2D) for solvent-surface-solvent (SSS) events
point is marked as “inside protein” if there is at least one protein atom within 1.6 Å.
Next, the solvent excluded surface is calculated using the Connolly algorithm
(Connolly
1983
) and the surface vertices' coordinates are stored. In the Connolly
algorithm, a hypothetical probe sphere (usual radius 1.4 Å) rolls over the protein.
The Connolly surface is a combination of the van der Waals surface of the protein
and the probe spheres surface, if the probe is in contact with more than one atom.
A grid point is marked as “near surface” if a surface vertex is within 1.0 Å. All the
other grid points are marked as “in solvent”. A sequence of grid points, which starts
and ends with “near surface” grid points and which has “in solvent” grid points in
between, is called a surface-solvent-surface (SSS) event. LIGSITE
csc
scans seven
directions, the x, y, z directions and four cubic diagonals, for such SSS events. If the
number of surface-solvent-surface events of a solvent grid exceeds a minimal
threshold (MINSSS, 5 in this work), then this grid is marked as pocket. Finally, all
pocket grid points are clustered according to their spatial proximity. I.e. if a pocket
grid point is within 3 Å to a pocket grid point cluster, it is added to this cluster.
Otherwise, it becomes a new cluster. Next, the clusters are ranked by the number of
grid points in the cluster. The top three clusters are retained and their centres of
mass are used to represent the predicted pocket sites. This first extension to the basic
LIGSITE algorithm is called LIGSITE
cs
. For LIGSITE
csc
, the top three pocket sites
are re-ranked according to the degree of conservation of the involved surface residues
around the pocket sites. To be precise, the conservation score is the average conservation
of all residues within a sphere of certain radius (8 Å here) of the centre of mass of
the cluster. The conservation score for each residue in a given PDB ID is obtained
from the ConSurf-HSSP database (Glaser et al.
2005
) , ranging from 1 (less conserved)


















































































































Search WWH ::

Custom Search