Biology Reference
In-Depth Information
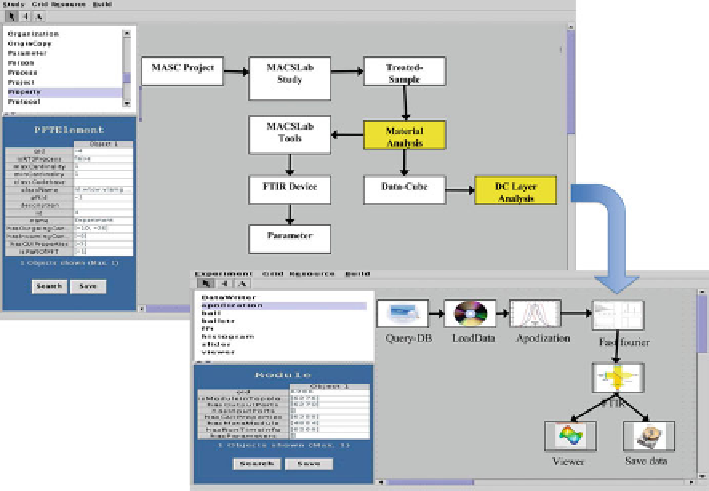
Fig. 7.5
This excerpt from the MACS Lab PFT shows both metadata descriptions (
white boxes
)
and automated experiment steps executed on the distributed infrastructure (
grey boxes
). The inset
shows an associated experiment work fl ow for DC analysis
information about the painting, sampling spot and device parameters. The PFT,
together with the descriptive fields, is stored in a database and can be subject to
queries (Hendrikse et al.
2003
) .
PFTs offer a clear separation between the information needed for at the design
phase of an experiment and the information needed at the execution time. Such
separation will reduce the amount of information the scientists have to deal with for
large-scale experiments. A similar abstraction also exists in Pegasus (Deelman et al.
2004
). The main difference between these abstract workflows and PFTs is that
Pegasus workflows can be made executable by mapping them to the underlying
resources, while PFTs provide the context of the experiment. PFTs have the advan-
tage of being decoupled from the execution of the experiments and, consequently,
can be used to point to experiments executed in different workflow systems.
7.5.2
Ontology-Based Approach
Experience has shown that a successful e-Science framework has to abstract the
scientists from all information technology issues. In the first approach to PFT a
database expert is needed to define the required underlying data structure. A more
practical approach will be to allow domain experts to describe the PFTs at a very high
Search WWH ::

Custom Search