Biology Reference
In-Depth Information
The requirement for local unfolding in order for perox-
iredoxin to complete its catalytic cycle in the detoxifica-
tion of hydrogen peroxide is very interesting and may
be a general mechanism used by other redox factors.
A
PE1 AS A REDOX FACTO
R
Since the initial discovery of APE1's (Ref-1) redox
activity in regulating the DNA-binding activity of
AP-1,
24,82
APE1 has been reported to reduce a number
of ubiquitous (i.e. AP-1, Egr-1, NF-
k
B, p53, CREB, HIF-
1
a
) or tissue-specific transcription factors (i.e. PEBP-2,
Pax-5 and -8, TTF-1)
24,67,68,83
e
89
APE1 is a multifunc-
tional protein that has apurinic/apyrimidinic endonu-
clease activity essential for BER, redox activity,
transcriptional regulatory activity,
90
and most recently
RNA-cleavage activity;
91,92
however, it is APE1's redox
activity that plays an important role in regulating the
expression of a large number of DNA repair proteins.
Evolution of Redox Activity in APE1
Although APE1 is reported to have distinct
N-terminal redox and C-terminal repair domains,
93
these functional domains are not independently folded
domains within the protein, i.e. structural domains
(
Figure 11.6
). Furthermore, the repair and redox activi-
ties are not coordinated within human APE1. The AP
endonuclease activity of APE1 is conserved from
bacteria to man, while the redox function is unique to
mammals. Thus, APE1 and E. coli exonuclease III, the
major AP endonuclease found within E. coli, are closely
related in terms of structure (r.m.s.d. 1.5
˚
), retaining
not only the same overall fold and topology but also
very similar endonuclease active sites.
9
The sequence
identity between APE1 and exonuclease III is ~28%.
The most obvious structural difference between human
APE1 and exonuclease III is an additional 62 N-terminal
residues found only in APE1. Within this N-terminal
region of APE1 is a nuclear localization sequence as
well as a nucleolar localization sequence that binds
directly to nucleophosmin.
91,94
However, addition of
N-terminal residues alone does not confer redox
activity; zebrafish APE includes a similar N-terminal
addition but lacks redox activity.
47
The question of which Cys residues are required for
APE1's redox activity has not yet been fully answered
and continues to be a source of controversy in the liter-
ature. In contrast to molecules such as TRX and GRX,
which maintain the general redox status of the cell,
APE1 does not contain two Cys residues within a C-X-
X-C motif. Thus, the mechanism by which APE1 reduces
transcription factors is likely to differ from that of thio-
redoxin or glutaredoxin. Of the seven Cys residues
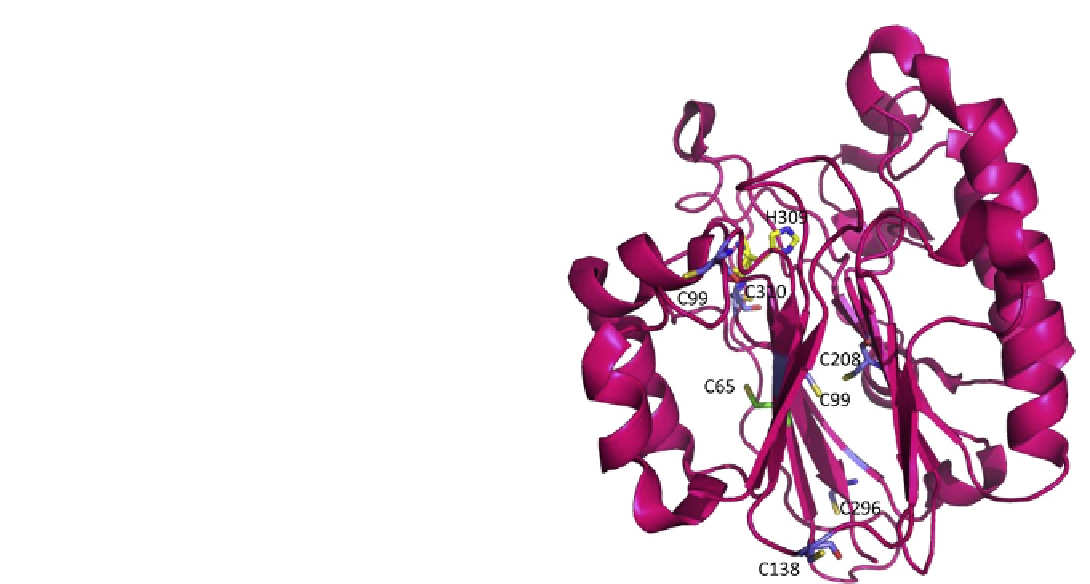
FIGURE 11.6
Apurinic/apyrimidinic endonuclease (APE1),
shown as a pink ribbon rendering, has seven Cys residues, 65, 93, 99,
138, 208, 296, 310, shown as stick renderings. Cys 65 is shown with
carbon atoms in green, oxygens, red, and sulfur, yellow. All other Cys
residues are shown with carbon atoms in blue, oxygen, red, and
sulfur, yellow. The redox active Cys residues 65 and 93 are located on
opposite sides of the beta sheet in which they are found. No disulfide
bonds are present in the crystal structures reported for APE1. His 309,
a critical active site residue, is shown as a stick model with carbon
atoms in yellow, oxygen in red, and nitrogen in blue. (Please refer to
color plate section).
(65, 93, 99, 138, 208, 296, 310) present in hAPE1, Cys 65
was identified as the critical residue required for redox
activity through analysis of single Cys to Ala substitu-
tions within APE1 (see
Figure 11.6
).
95
Investigation of
the role of Cys residues within APE1 was based on the
finding that a Cys residue within the DNA-binding
domain of the transcription factor c-Jun was subject to
oxidation leading to loss of DNA-binding and was
reduced by APE1.
23,24,82
Subsequently, the crystal struc-
ture of human APE1 was reported,
96
revealing that Cys
65, a residue unique to mammalian sequences, is
a buried residue located on the first beta strand in the
fold, which is part of a beta sheet in the core of the pro-
tein (
Figure 11.6
). The residue equivalent to the hAPE1
Cys 65 in exonuclease III based on structural alignment
is Val 4 while that in zebrafish APE1 (zApe) is Thr 58.
47
Only two Cys residues, Cys 208 and Cys 310, are
conserved in both hAPE1 and E. coli exonuclease III,
but within vertebrate APEs, all Cys residues except
Cys 65 and Cys138 are conserved.
47
The structures of
zAPE and human APE1 are quite similar suggesting
that vertebrate APEs are highly conserved. In fact,