Biology Reference
In-Depth Information
domain (reviewed in
65,129
)(
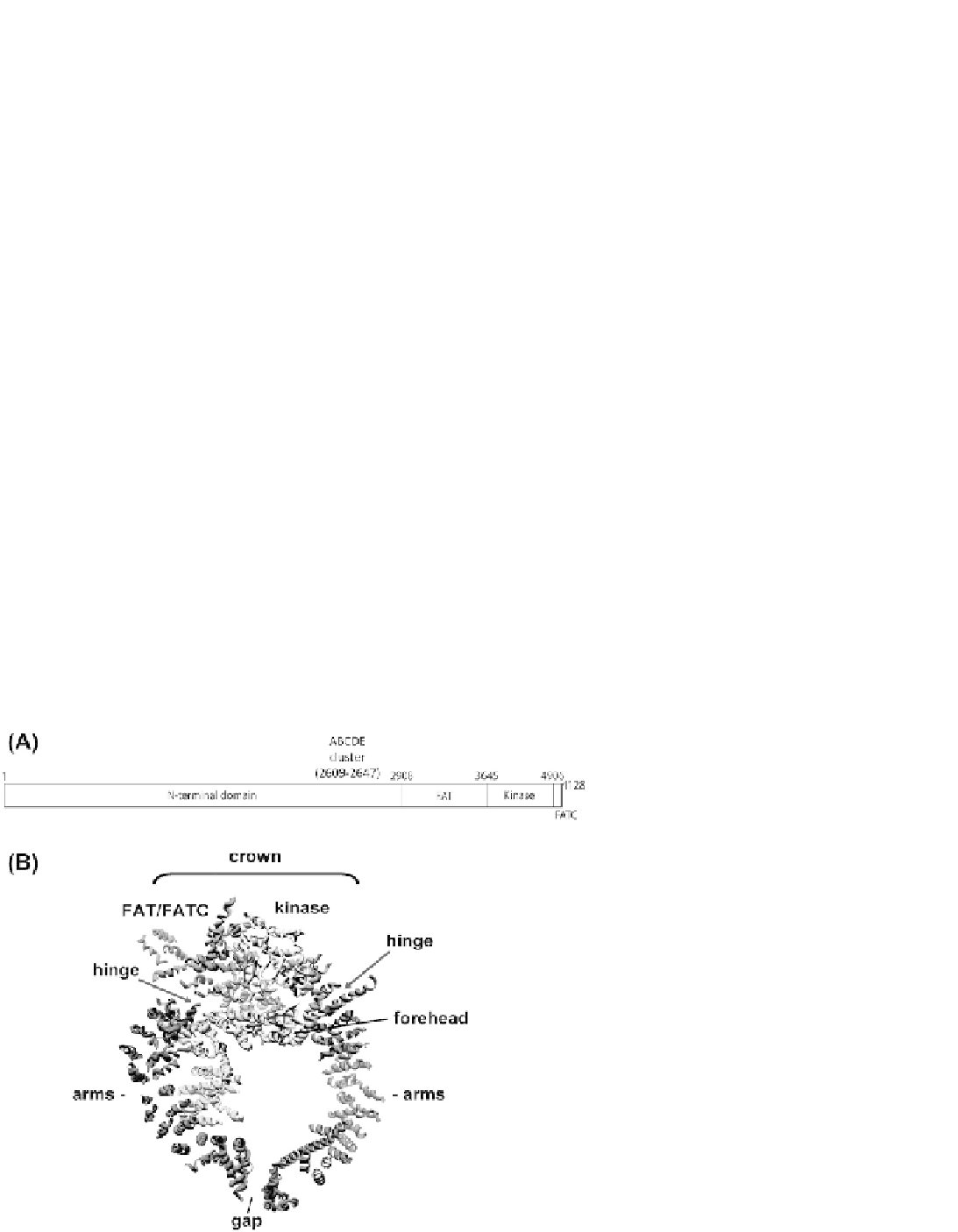
Figure 8.7A
). The recent
low resolution X-ray structure of DNA-PKcs reveals
a horseshoe shaped structure where the FAT domain
forms the arms of a pair of pincers with the FAT-
kinase-FATC domains located at the apex, forming the
head or crown
137
(
Figure 8.7B
). The arms of the pincers
(corresponding to the N-terminal
a
-helical domain)
surround a large central channel that likely accommo-
dates dsDNA.
138,139
Interestingly, the X-ray structure
also revealed a gap at the base of the pincers, and the
region connecting the arms of the pincers to the crown
domain (labeled hinge regions in
Figure 8.7B
)is
proposed to be highly flexible, suggesting that confor-
mational changes regulate the positions of the pincer
arms and the dimensions of the gap.
137
DNA-PKcs is a member of the PIKK family of serine
threonine protein kinases
78
and, like the related protein
kinase ATM preferentially phosphorylates peptide
substrates on serines and threonines that are followed
by glutamine (SQ/TQ motifs).
79
Also, like ATM, DNA-
PK activity is inhibited by wortmannin.
98
The protein
kinase activity of DNA-PKcs is essential for
NHEJ
140,141
and small molecule inhibitors of DNA-PK
radiosensitize cells,
142
making DNA-PKcs a possible
therapeutic target
143
(discussed below).
DNA-PK has robust
in vitro
protein kinase activity
and numerous
in vitro
substrates have been identified.
Although combinatorial peptide libraries have revealed
that the optimal consensus is SQ/TQ (
79
), and many
in
vitro
substrates are phosphorylated on SQ/TQ motifs,
DNA-PKcs also phosphorylates other motifs, such as
serines followed by leucine (see for example,
144,145
).
For example, hnRNPU is phosphorylated by DNA-PK
in vitro
on a non-SQ/TQ site and is phosphorylated
in
vivo
in a DNA damage and DNA-PK dependent manner
suggesting that DNA-PK targets both SQ/TQ and non-
SQ/TQ sites
in vivo.
146,147
Given that DNA-PKcs' kinase activity is required for
NHEJ a logical question is whether DNA-PK-mediated
phosphorylation of the core NHEJ proteins is required
for DSB repair. However, although DNA-PKcs phos-
phorylates Ku70, Ku80,
145
XRCC4,
144
XLF,
148
DNA
ligase IV,
149
and Artemis
150
in vitro
, to date there is little
evidence that any of these phosphorylation events is
required for DSB repair
in vivo
(reviewed in
65,151
).
Indeed, mutation of multiple
in vitro
DNA-PK phos-
phorylation sites in Ku, XRCC4, XLF and Artemis does
not affect radiation sensitivity or V(D)J recombination.
Moreover, the majority of
in vitro
DNA-PK phosphoryla-
tion sites in XRCC4, XLF, and Artemis are located in
disordered C-terminal regions of the proteins that are
not required for NHEJ or V(D)J recombination.
To date, the only clearly identified physiological target
of DNA-PK kinase activity is DNA-PKcs itself. DNA-
PKcs undergoes extensive autophosphorylation
in vitro
which correlates with loss of protein kinase activity
and dissociation of autophosphorylated DNA-PKcs
from DNA-bound Ku.
134,152
e
155
Together, these studies
suggest that autophosphorylation promotes release
of DNA-PKcs from DNA-bound Ku (reviewed in
65
).
FIGURE 8.7
Domains and low resolution struc-
ture of DNA-PKcs. (A) Domains of DNA-PKcs: FAT,
FRAP (FKBP12-rapamycin-associated protein),
ATM, TRRAP (transactivation/transformation-
domain-associated protein) domain; FATC, FAT C-
terminal domain. Amino acid numbers are indi-
cated on the top of the figures. The location of the
ABCDE/Thr 2609 cluster of autophosphorylation
sites (amino acids 2609-2647) is also shown. See text
and Mahaney
et al.
, 2009
65
for details. Additional
phosphorylation sites are described by Dobbs
et al.
,
2010.
162
(B) The X-ray structure of DNA-PKcs at 6.6
˚
as determined by Sibanda
et al.
, 2010.
137
From
Dobbs
et al.
, 2010.
162