Biology Reference
In-Depth Information
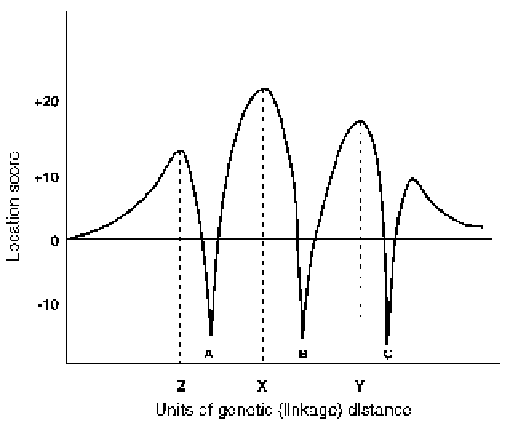
FIGURE 3.2. Multipoint linkage analysis. A, B and C represent the known linkage
relationships of three polymorphic marker loci. X, Y and Z represent, in descend-
ing order of likelihood, the probable position of the disease locus. (Reprinted from
Emery's Elements of Medical Genetics, 10th ed, Meuller, RF, Fig. 3.2, Copyright
1998, by permission of Churchill Livingston.)
In the case of autosomal dominant nonsyndromic sensorineural hearing
impairment, single large families have been collected in which there are
affected individuals in multiple generations in order to overcome this
problem. An example of this approach was the analysis of the large Puerto
Rican family with autosomal dominant low-frequency hearing impairment
(Leon et al. 1981), which led to the identification of the first locus for auto-
somal dominant nonsyndromic sensorineural hearing impairment,
DFNA1
,
on the long arm of chromosome 5 (Leon et al. 1992).
In the case of autosomal recessive nonsyndromic sensorineural hearing
impairment, the problem is potentially even more difficult. In the majority
of families with autosomal recessive nonsyndromic sensorineural hearing
impairment, there are usually at most two or three affected siblings. Pooling
the Lod scores from these families is very unlikely to demonstrate linkage.
This problem has been overcome by use of the method of autozygosity
mapping, which involves using samples from families in which the parents
of affected offspring are consanguineous (Lander and Botstein 1987).
Affected individuals in such pedigrees will have regions of their genome
that are homozygous for polymorphic DNA markers because they were
inherited from a common ancestor (Fig. 3.3). Identification of large con-
sanguineous families from ethnic isolates means a single family can be
sufficient to establish linkage (Mueller and Bishop 1993; Kruglyak et al.