Information Technology Reference
In-Depth Information
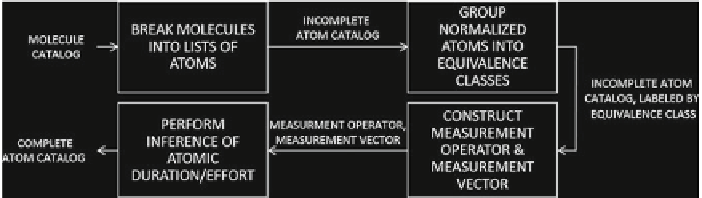
Fig. 16.10
Method for estimating the time durations of atomic steps in a recipe
4. If the measurement operator is sufficiently invertible, proceed to Step 5; otherwise
return to Step 2 and categorize into coarser equivalence classes,
5. The measurement operator and the measurement vector are used as inputs to an
inference algorithm that solves the inverse problem to find the durations of each
atom.
Since we want to determine how much time it takes to perform granularized
atomic tasks when we only have access to the duration of molecules composed of
several atomic tasks all mixed together, several difficulties must be overcome. First,
we need to ensure the inverse problem to be solved is not “undercomplete” (more
equations than unknowns) by grouping atomic tasks into equivalence classes whose
members will be treated as having the same duration as any other member of the
class. Second, we need to normalize/rescale duration to take into account disparities
in quantities, tools, etc. Finally, we need to develop an inference algorithm to find the
unknown atomic durations from the known durations of molecules and the known
memberships of atoms in molecules.
Time is an extensive quantity and essentially adds up linearly. Letting
y
be the
total time of the molecular piece of work, and
x
a
,
x
b
,
x
c
be the times for three atomic
steps, we assume that
y
x
c
. We also assume that there is a finite set
of N possible recipe steps from which these steps are chosen. We can therefore use
indicator variables
a
i
to write a generalized expression for the sum, where
a
i
=
=
x
a
+
x
b
+
0
for absent steps and
a
i
=
1 for present steps:
⊡
⊣
⊤
⊦
=
x
1
x
N
a
1
···
a
N
y
,
where the
are unknown and to be inferred. We
see that using data on just a single molecule, the inference problem is undercomplete.
Now consider a catalog with many molecules
M
, with the number of possible recipes
steps denoted as
N
:
{
a
i
}
and
y
are known whereas the
{
x
i
}
Search WWH ::

Custom Search