Environmental Engineering Reference
In-Depth Information
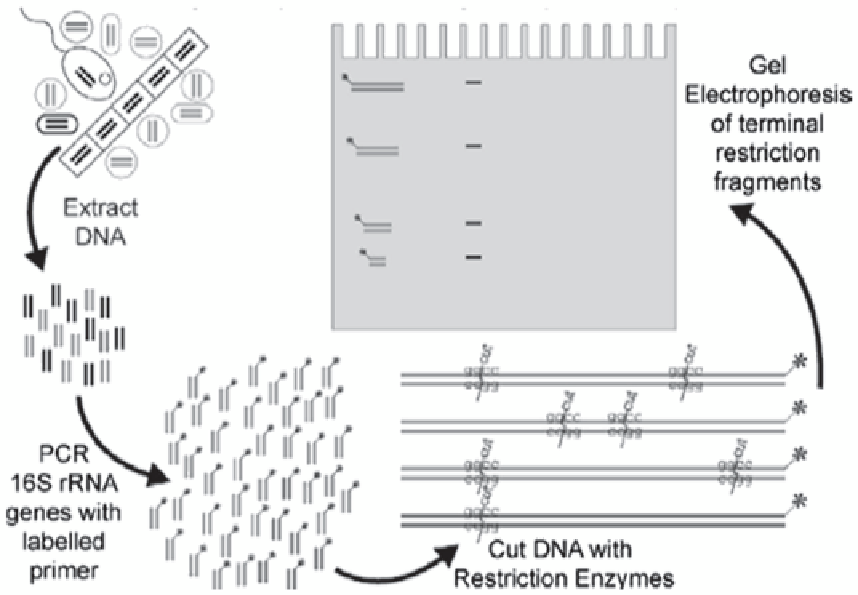
Fig. 4.4
Diagrammatic representation showing T-RFLP.
Mixed population is amplified using a
16S
primer with a
fluorescent tag.
PCR
product is cut with a 4 bp cutting re-
striction endonuclease. Different sequences will give dif-
ferent length fragments. Sample is injected into a capillary
sequencer to sort fragments by size
analysis (ARDRA) and provides an alternative
method for rapid analysis of microbial commu-
nity diversity in various environments. It follows
the same principle as RFLP except that one PCR
primer is labeled with a fluorescent dye (Fig.
4.4
).
Similar in principle to RFLP and T-RFLP, ri-
bosomal RNA (rRNA) intergenic spacer analysis
(RISA), automated ribosomal RNA (rRNA) in-
tergenic spacer analysis (ARISA), and ARDRA
provide ribosomal-based fingerprinting of the
microbial community. In RISA and ARISA, the
intergenic spacer region between the 16S and
23S ribosomal subunits is amplified by PCR,
denatured and separated on a polyacrlyamide gel
under denaturing conditions. This region may en-
code tRNAs and is useful for differentiating be-
tween bacterial strains and closely related species
because of heterogeneity of the intergenic space
length and sequence (Scheinert et al.
1996
). Se-
quence polymorphisms are generally detected by
silver staining in RISA. In ARISA, as the name
suggests fluorescently labeled forward primer is
detected automatically (Fisher and Triplett
1999
).
Both methods can provide highly reproducible
bacterial community profiles. The process of
RISA requires large quantities of DNA, relative-
ly longer time requirement, insensitivity of silver
staining in some cases, and low resolution which
can be taken as its limitations. ARISA has better
sensitivity than RISA and is less time consum-
ing but traditional limitations of PCR also applies
for ARISA (Brown and Fuhrmans
2005
). RISA
has been used to compare microbial diversity in
soil, in the rhizosphere of plants (Borneman and
Triplett
1997
), and in contaminated soil (Ranjard
et al.
2000
).
Presently, DNA-DNA hybridization has been
used along with DNA microarrays to detect and
identify bacterial species or to evaluate microbial
diversity (Rastogi and Sani
2011
). This tool could
Search WWH ::

Custom Search