Environmental Engineering Reference
In-Depth Information
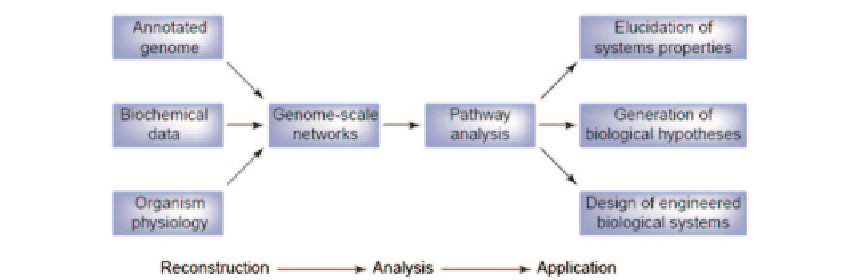
Fig. 3.1
An overview of pathway analysis. (Reproduced from http://www2.bio.ifi.lmu.de/lehre/SS2007/SEM_Ad-
vanced/Folien/Extreme_pathways.pdf)
things must work together at many different lev-
els—from organs to cells to genes. Cells are con-
stantly receiving cues from both inside and outside
the body, which are prompted by such things as
injury, infection, stress or even food. To react and
adjust to these cues, cells send and receive signals
through biological pathways. The molecules that
make up biological pathways interact with signals,
as well as with each other, to carry out their chosen
tasks. Biological pathways can also produce small
or large outcomes. Scientists are researching that
biological pathways are far more complex than
once believed. Most pathways do not start at point
X and end at point Y. In fact, many pathways have
no real restrictions, and they often work together
to complete tasks. When multiple biological path-
ways interact with each other, it is known as a bio-
logical network (Fig.
3.1
). These pathways have
then been grouped conceptually as functional units
such as glycolysis or the tricarboxylic acid cycle
(TCA) cycle. This type of pathway definition is
useful for identifying portions of the metabolic
network, but the divisions are somewhat vague
between the point where one pathway ends and
another begins. In addition, this type of pathway
definition does not relate to the overall functions
of the network as a whole.
Clarke (Fig.
3.2
). The theory was developed to
study instability in inorganic chemical networks.
This was the first attempt to apply convex analy-
sis to reaction networks but it was never extended
to living systems. This was followed by some
work using AI to search through reaction net-
works following along the lines of graph theory
and was taken another step further by Mavro with
the introduction of stoichiometric constraints.
Both of these approaches lacked a sound theo-
retical basis. In 1994, Schuster became the first
to apply convex analysis to metabolic networks
with the introduction of a non-unique set of el-
ementary modes. This theory was applied a few
years later by Liao to optimize bacterial strain de-
sign for the high-efficient production of aromatic
amino acids. So at this point in time, pathway
analysis was just beginning to be applied but still
lacked a unified theoretical foundation, which is
where the present work comes in.
3.2
Extreme Pathways
3.2.1
Definition
Extreme pathways are defined as vectors de-
rived mathematically and can be used to char-
acterize the phenotypic potential of a defined
metabolic network (Schilling et al.
1999
,
2000
).
Extreme pathway analysis has the following
characteristics:
3.1.2
A Brief History of the Field
of Pathway Analysis
The first work on pathways can be traced back
to 1980 with the development of SNA by Bruce
Search WWH ::

Custom Search