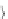Database Reference
In-Depth Information
C_Id
Original
Customer Sequence
Transformed
Customer Sequence
After
Mapping
1
(30) (90)
(30)
(90)
{1}{ 5}
2
(10 20) (30) (40 60 70)
(30)
(40) , (70) , (40 70)
{1}{ 2, 3, 4}
3
(30 50 70)
(30) , (70)
{1, 3}
4
(30) (40 70) (90)
(30)
(40) , (70) , (40 70)
(90)
{1}{ 2, 3, 4}{ 5}
5
(90)
(90)
{5}
Fig. 11.1
Transformed Database. (Adapted from [
8
])
The inefficiency of AprioriAll algorithm mainly stems from its computationally
expensive data transformation phase which transforms each transaction to a set of
litemsets (frequent itemsets) in order to find sequential patterns.
3.1.2
GSP
The same authors later proposed a new algorithm called GSP [
9
], which overcomes
the drawbacks of AprioriAll and extends it by allowing time constraints, sliding time
windows, and taxonomies. GSP is also a horizontal data format based sequential
pattern mining algorithm. Based on the downward closure property of a sequen-
tial pattern, GSP adopts a multiple-pass, candidate-generation-and-test approach in
sequential pattern mining.
The algorithm is outlined as follows. The first scan finds all of the frequent items
which form the set of single item frequent sequences. Each subsequent pass starts
with a seed set of sequential patterns, which is the set of sequential patterns found
in the previous pass. This seed set is used to generate new potential patterns via a
join phase, called candidate sequences. This join step is done in the following way.
A sequence
s
1
joins with
s
2
if the subsequence obtained by dropping the first item
of
s
1
is the same as the subsequence obtained by dropping the last item of
s
2
. The
candidate sequence generated by joining
s
1
with
s
2
is the sequence
s
1
extended with
the last item in
s
2
. The added item becomes a separate element if it is a separate
element in
s
2
, and part of the last element of
s
1
otherwise.
Thus, each candidate sequence contains one more item than a seed sequential
pattern, where each element in the pattern may contain one or multiple items. The
number of items in a sequence is called the length of the sequence. So, all the
candidate sequences in a pass will have the same length. The scan of the database
in one pass finds the support for each candidate sequence. All of the candidates
whose support in the database is no less than
min_support
form the set of the newly
found sequential patterns. This set then becomes the seed set for the next pass.
The algorithm terminates when no new sequential pattern is found in a pass, or no
candidate sequence can be generated.
The method is illustrated using the following example [
3
].
Example
1
(GSP)
Given
the
sequence
database
D
in
Table
11.1
and
min
_
suppor t
2, GSP first scans
D
, collects the support for each item, and
finds the set of frequent items, i.e., frequent length-1 subsequences (in the form of
=