what-when-how
In Depth Tutorials and Information
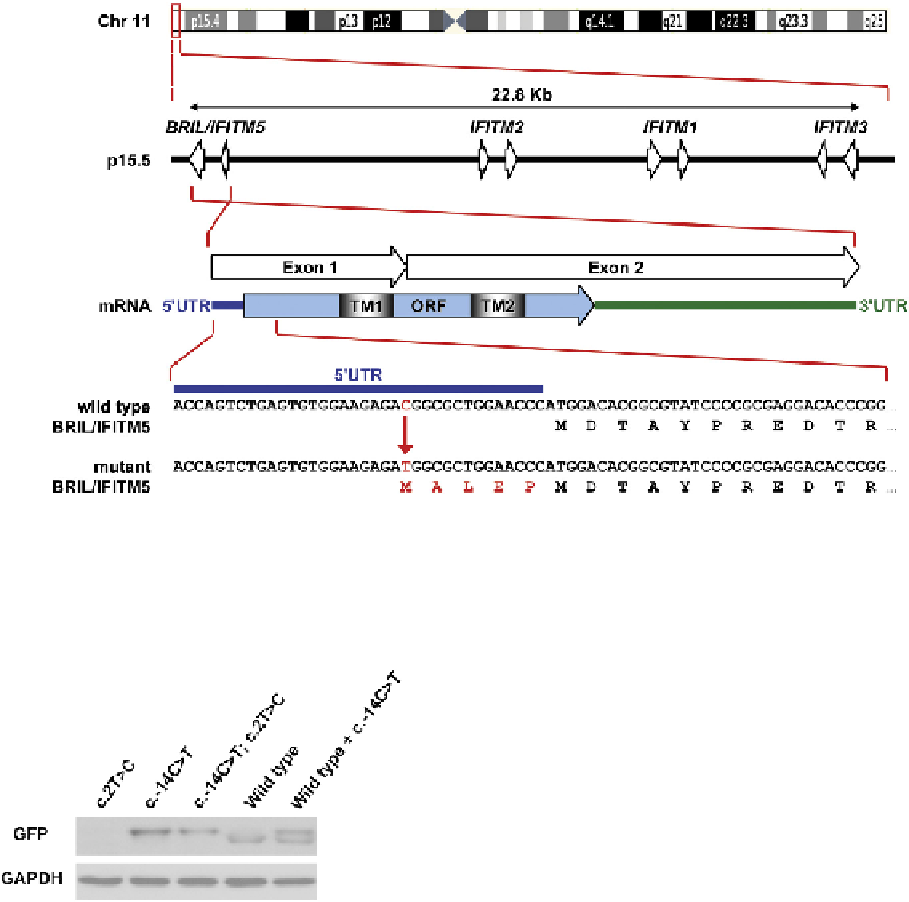
FIGURE 20.1
Localization and architecture of the
BRIL/IFITM5
on chromosome 11 and encoded protein.
BRIL
and other
IFITM
genes are
clustered within a 22.8 Kb segment at p15.5. All members have two coding exons represented by arrows. Normal
BRIL
transcript encodes a 132
amino acid protein having two transmembrane domains (TM). The mutation found in a type V OI patient (c.-14C>T) indicated by the red arrow
is present in the 5′ untranslated region of the gene (blue line). The additional five amino acids generated by this mutation are shown under the
gene sequence.
domains. Immunolocalization studies using tagged BRIL
10
and IFITM3
17,18
confirmed that they have both their N-
and C-termini extruding out into the extracellular space
(
Figure 20.3
), although this predicted model has been chal-
lenged recently, at least for IFITM3.
19,20
Furthermore, other
members like IFITM3 also seem to localize preferentially
into the endosome compartment,
19,20
while BRIL localizes
to plasma membranes.
10
BRIL presents some other fea-
tures which make it a distinctive member. For instance,
unlike other
IFITM
members which are highly inducible
by type I interferons (α and β) due to the presence of inter-
feron regulatory elements in their promoter region,
BRIL
transcription is not responsive to interferons.
21
Rather,
ongoing experiments in our laboratory indicate that regu-
lation of
BRIL
transcription is mainly activated by Sp1/
Sp3 and Gli transcription factors.
44
Expression of an anti-
sense transcript covering the neighboring
Ifitm2
gene and
part of the 3′ untranslated region of
Bril
was also shown to
positively impact on
Bril
expression in rat osteoblast cul-
tures.
22
Whether this mechanism of regulation is operative
in human cells is unknown. More importantly, expression
of
BRIL
is mostly restricted to osteoblasts whereas other
IFITMs
are ubiquitous. We have shown by
in situ
hybrid-
ization, RNA profiling and immunohistochemistry that
in mouse and rat,
Bril
is mostly restricted to osteoblasts.
10
Expression was also shown to be enriched in bone tis-
sues in the human
10
and the tammar wallaby.
16
A separate
study also found increased
Bril
expression under culture
conditions favoring an osteogenic behavior.
23
FIGURE 20.2
Transient expression assay showing proteins
produced by wild-type and mutant constructs. When the original
start codon was mutated (c.2T>C), no translation was observed.
Translation started at the start codon generated by the mutation
(c.-14C>T) regardless of the original start codon status. Its translation
product was slightly larger than that of the wild-type. Co-transfection
with wild-type and c.-14C>T mutated constructs produced double
bands.
(Reproduced from
11
with permission.)
and 10).
15,16
The mouse has two other members (IFITM6
and 7). The term “dispanin” has been coined recently to
encompass IFITMs into an even larger family of proteins
that have two transmembrane passages.
15
The classification of
BRIL
as part of this family is
mainly based on generally similar characteristics as
depicted in
Figure 20.1
.
BRIL
,
IFITM1
,
2
and
3
are all clus-
tered within 25 kb on chromosome 11, they all possess a
similar gene architecture comprising two small coding
exons separated by a short intron, and potentially similar
predicted protein topology having two transmembrane