TABLE 9.2 Viroids
Family/genera
Type species
Genome size
Host(s)
Comments
Popsiviroidaea
Popsiviroid
PSTVd
356 to 375 nt
Presence of central conserved region and lack of self-cleavage
Plants
mediated by hammerhead ribozyme; replicate by an
Hostuviroid
HpSVd
295 to 303 nt
asymmetric rolling circle strategy in nucleus of infected
Cocaviroid
CCCVd
246 to 301 nt
cells, probably using RNA polymerase II
Apscaviroid
ASSVd
306 to 369 nt
Coleviroid
CbVd-1
248 to 361 nt
Avsunviroidaeb
Avsunviroid
ASBVd
246 to 250 nt
Plants
Lack central conserved region; replicate by a symmetric
strategy in chloroplasts of infected plants using
Pelamoviroid
PLMVd
337 to 399 nt
chloroplastic RNA polymerase; can form self-cleaving
hammerhead ribozymes in both plus and minus strands
a
Formerly known as the Group B viroids.
b
Formerly known as the Group A viroids.
infect plants. However, hepatitis δ, which infects humans,
virulence of the viroid on infection of its plant host. For non-
has many viroid-like properties and may be related to viroids.
self-cleaving viroids, it is assumed that the concatenated
Many viroids are important agricultural pathogens, whereas
RNAs are cleaved and ligated by host-cell enzymes.
others replicate without causing symptoms. Viroids are
The self-cleaving viroids possess a hammerhead
often transmitted through vegetative propagation of plants,
ribozyme structure, illustrated in Fig. 9.5. The ribozyme
but can also be transmitted during agricultural or horticul-
activity cleaves the concatenated RNA at the points
tural practices in which contaminated instruments are used.
indicated by the arrows and ligates the ends to form cir-
Some viroids can be transmitted through seeds and at least
cular molecules. The viroid RNA is very compact in its
one viroid is transmitted by an aphid.
structure, with extensive secondary structure, including
On infection of a plant cell, viroid RNA is transported
pseudoknots.
to the nucleus. The circular RNA appears to be copied
There also exist a large number of satellites called viru-
by host-cell RNA polymerase II, using a rolling circle
soids. Virusoid RNAs are about 350 nt in length, and the
mechanism in which multimeric antigenomic sense RNA
RNA is a single-stranded, covalently closed circle. The
molecules are produced. The multimeric antigenome
mechanisms by which virusoid RNA is replicated have not
sense RNA can then be used as a template to make mul-
been precisely determined, but they appear to be viroid-
timeric genomic sense RNA. This synthesis may also
like and may replicate by the same mechanisms as viroids.
be performed by RNA polymerase II. The concatenated
At least some virusoid RNAs are capable of self-cleavage.
RNAs are cleaved and cyclized to produce the progeny
Virusoid RNAs are encapsidated by the capsid protein of the
viroid RNA molecules. In an infected cell, as many as
helper virus of which the virus is a satellite. Thus, transmis-
104 viroid RNAs can accumulate, most of them in the
sion occurs by conventional virus-like means, and virusoids
nucleus.
may have arisen from viroids that evolved a mechanism for
Some viroids are capable of self-cleavage by the con-
packaging using a helper virus.
catenated RNAs to produce genome-length RNAs, fol-
lowed by self-ligation to cyclize the unit-length molecule.
HEPATITIS δ
Other viroids are not capable of self-cleavage and ligation.
There are five groups of non-self-cleaving viroids, classi-
The hepatitis delta (δ) agent or virus (HDV) is a satellite
fied by the sequences in the central conserved region. The
structures of these five groups are shown in Fig. 9.4. The
of hepatitis B virus (HBV). It has a worldwide distribution,
conserved domains highlighted in the figure are thought to
although strains isolated from different regions of the world
be important for the replication of the viroid (i.e., to form
differ by up to 40% in their nucleotide sequence. The distri-
promoters recognized by RNA polymerase II) and for its
bution of HDV is not uniform around the world. Regions of
cleavage to produce unit-length molecules. A pathogenesis
particularly high prevalence include the Mediterranean basin,
domain is also highlighted. Changes in this domain affect the
the Middle East, Central Asia, West Africa, the Amazon
A.
B.
PSTVd
G GA
AA AC
CGCUUCAG
UCC CCGGGG
CNNGNGGUUCCUGUGGG
CUGGAGCG
AGG UGGCCCA
CGAAGUCA
ACAA
U CA
C G
U
C
G
C
C G
CG
U A
HSVd
A
A
U GGGG AA
A GCC CCGG GGCAACU C
G
A
A GG
G C
C
U CCCC
CGGU GGCC CG
AG
CCA
G C
CU A
AG
C
A U
C G
U G
UA
CCCVd
C G
A
G AA A
U GGGG A
GG
GAG
AUCC CCGGG
C
U CCCC
UAGGU GGCCCA
CUCA
AC A
UC A
PSTVd
ASSVd
UCGUC G UCGAC G A A GG
CNNGNGGUUCCUGUGGG
CAG
AGCUG
CC
GCG
AUCG
CbVd1
U
A
A
CUGGGU CCCU G GCAG CGCUGCA CGGA U
CNNGNGGUUCCUGUGGG
GGA
CGUU GCG
AA
A
Conserved Sequences
Domains
Terminal Left (TL)
Pathogenesis (P)
Terminal Conserved Region (TCR)
Terminal Conserved Hairpin (TCH)
Terminal Right (TR)
Variable (V)
Central Conserved Region (CCR)
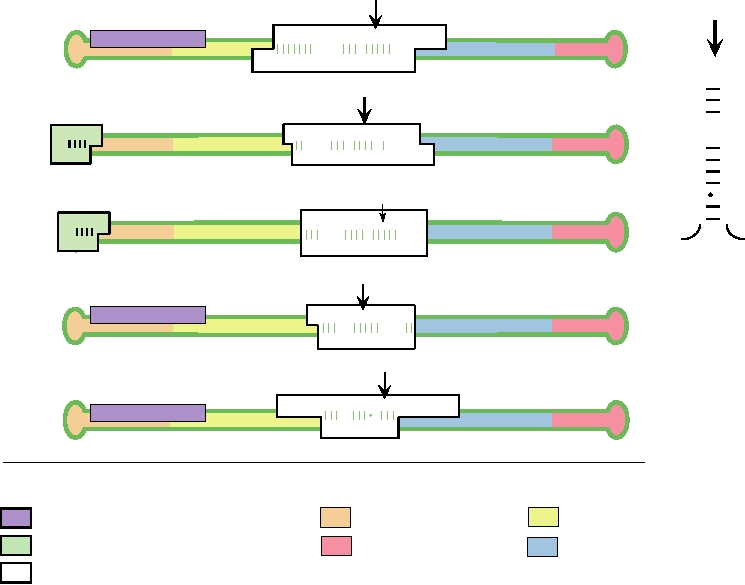
FIGURE 9.4 Models for the genomes of the type species of the five genera of non-self-cleaving viroids. They are as
follows: Popsiviroid: PSTVd, potato spindle tuber viroid; Hostuviroid: HSVd, hop stunt viroid; Cocadviroid: CCCVd,
coconut cadang-cadang viroid; Apscaviroid: ASSVd, apple scar skin viroid; Coleviroid: CbVd-1, Coleus blumei viroid-
1. (A) The RNA strand is shown as a green closed loop. Four functional domains (TL, TR, P, and V) are indicated with
different colors of shading. Three conserved sequences are boxed. The central conserved region (CCR) is a white box,
the terminal conserved region (TCR) is a lavender box, and the terminal conserved hairpin (TCH) is a green box. The
nucleotides in the upper strand of the CCR (dark green) can in each case form a stable stem and loop structure with the
top of the loop at the black arrow, as shown for PSTVd in (B). In this alternative configuration, the nucleotides that are
invariant within all five groups are shown in red. Adapted from Flores et al. (1997).
Basin, and certain islands in the South Pacific. HDV will
patients. The different outcomes following infection with
only replicate in cells that are simultaneously infected with
HDV are illustrated schematically in Fig. 9.7, in which the
HBV and its distribution is thus dependent in part upon the
symptomology at different times after infection is indicated.
distribution of HBV, which was shown in Fig. 6.29. However,
The illness caused by HDV is usually more serious than
as shown in Fig. 9.6, HDV is not uniformly distributed
that caused by HBV alone. The mortality rate from HDV
throughout the range of HBV. The percentage of hepatitis B
infection is 220%, 10-fold higher than the rate for HBV
patients that are also infected by infection with HDV ranges
infection alone, which is the next most severe form of viral
from 5% to more than 60% in different geographic areas.
hepatitis. Most cases of HDV infection are probably clini-
Infection of humans by HDV can either occur by simultane-
cally important. It is estimated that 460 million people in the
ous infection with both HBV and HDV, or by superinfection with
world are chronically infected with HBV of whom perhaps
HDV of a person who is chronically infected with HBV. In
20 million are also chronically infected by HDV. All persons
the case of coinfection, a chronic infection by HDV, which
chronically infected with HBV that are not already infected
requires that HBV also establish a chronic infection, is
by HDV are at risk for contracting HDV and suffering a
established only 13% of the time. Most often the infection
more severe form of hepatitis.
is completely resolved and recovery occurs. In contrast,
The mechanisms by which HDV is transmitted are not
superinfection of chronically infected HBV patients with
understood. It is conjectured that poor hygiene together with
HDV leads to chronic infection by HDV in 7080% of
intimate contact among people who are infected with the
A.
G
A
G
A
20
40
G
55 U
CA A
1
U
A
A
C G
C
C
G
C
C
AGU U U C G C U A U U C A A G GCU C A U C A G UG G C U U A G C CA G A C U U
A
285
C
U CA A A G C G G U A A GU U CU G A G U AG U C A C C G A A U C G GU CU G A G
339
A
C
U
A A
A
A
U
C
A
UUU
300
G
320
5
3
5
1
3
G
C
C.
B.
Minus strand
Plus strand
A
U
U
G
G
C
U
A
55
A 285
U
A
U
U
U
C
G
U
A
U
A
U
A
320
C
G
C
G
AA
U
AA
U
20
C
AU
G A G A C GA
U GU G C U
C
AU
A C G G C GA
U G GG C U
A
G
A
A
A
A
CA CA CGA
CU C UG A
UG C C G A
C A C C C GA
GU
GU
U
G
G
U
U AG
C AG
300
40
FIGURE 9.5 Predicted secondary structure of peach latent mosaic viroid (PLMV) (Family Avsunviroidae, Genus
Pelamoviroid) RNA in solution. (A) The entire viroid. For most of the molecule, only the backbone is shown, with
hydrogen bonds indicated by bars and GU pairs indicated with dots. The numbering of the nucleotides is arbitrary. The
structure is predicted to be even more compact due to pseudoknots formed by the regions joined by purple lines. The
nucleotides making up the minus-strand and plus-strand hammerhead ribozymes are shown in green and blue letters,
respectively. The nucleotides conserved in all hammerheads in Avsunviroidae are boxed and shaded, and the sites of
cleavage are marked by arrows. (B) and (C) The structures of the two hammerhead ribozymes, using the same color
conventions. The minus-strand hammerhead is made up of the complement of the sequence shown in green in (A).
Adapted from Flores et al. (1997) and Pelchat et al. (2000).
virus may be an important source of transmission in many
Replication of the HDV Genome and
parts of the world. In developed countries, contaminated
Synthesis of mRNA
blood products and sharing of needles by drug users are
The HDV genome is a single-stranded, covalently closed
important in the spread of the virus, but these are not impor-
circular RNA molecule of 1.7 kb. The HDV genome can
tant modes of transmission worldwide. Sexual transmission
be thought of as a viroid into which has been inserted a
may occur, but again this does not appear to be an important
gene encoding a single polypeptide, the hepatitis δ antigen
component of the transmission of the virus on a worldwide
(HDAg). As is the case for viroid RNA, HDV RNA has a
basis.
high degree of secondary structure, with about 70% of the
Since HDV depends on HBV for its propagation, control
molecule being base paired internally so that it forms a rod-
of HDV is dependent on control of HBV. Current HBV vac-
like structure. The HDV genome has minus-sense polarity,
cines are highly effective at preventing HBV infection and
that is, it is complementary to the sequence that is trans-
the increasing levels of vaccination against HBV, together
lated into the HDAg. The structures of the genomic RNA,
with increased screening for the presence of HBV in blood
the mRNA for the HDAg, and of the antigenomic RNA are
products, has resulted in a dramatic reduction in recent years
illustrated in Fig. 9.8A. The 0.8-kb mRNA is capped and
of new infections by HDV.
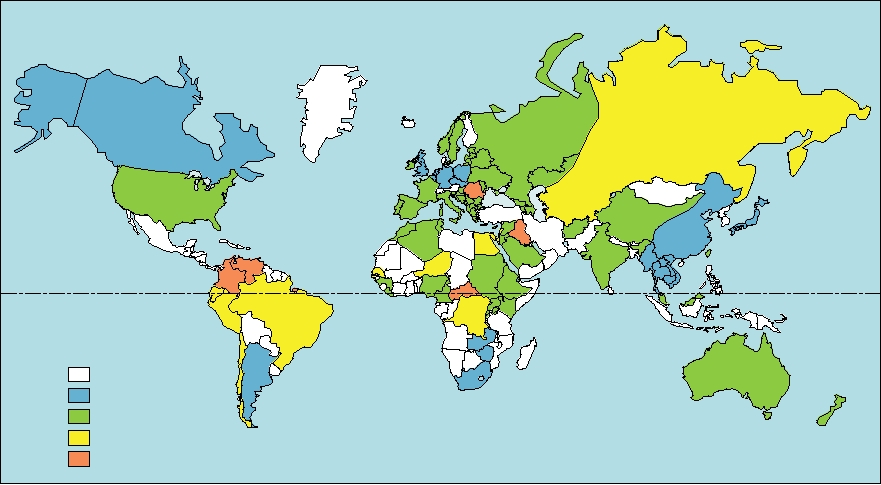
Kuwait
Equator
Percent of Hepatitis B Patients
with Hepatitis d Antigen
No Data
0-5%
6-20%
21-60%
>60%
FIGURE 9.6 Worldwide distribution of hepatitis δ infection as measured by the presence of hepatitis δ antigen in the
serum of hepatitis B infected patients with hepatitis. Adapted from Fields et al. (1996) p. 2826.
polyadenylated as is common for mRNAs. The 1.7-kb
unit-length molecules are then cyclized. Although HDV
antigenome is an exact complement of the genomic RNA
RNA is capable of self-ligation, this process appears to be
and, like genomic RNA, is also circular.
inefficient and it is thought that a cellular ligase is responsi-
Following infection by the agent, the RNA is transferred
ble for most cyclization of HDV RNA monomers.
to the nucleus. The HDAg, of which there are about 70
There are differences in the synthesis of genomic and
copies in the virion, is required for this. In the nucleus, the
antigenomic RNA, including differences in their rates of
RNA is replicated by mechanisms related to those used by
synthesis, different sensitivities of their synthesis to drugs,
viroid RNA. However, there is the added complication that
in the requirement for different forms of HDAg (described
an mRNA for HDAg must also be produced. Thus, there are
later) for genomic and antigenomic RNA synthesis, and
three elements to the replication of RNA: the production
in the transport of the RNAs to different places within the
of an antigenomic RNA template from genomic RNA, the
cell after synthesis. Furthermore, synthesis of genomic
production of mRNA for HDAg from genomic RNA, and
and antigenomic RNA may occur in different places in the
the production of genomic RNA from antigenomic RNA
nucleus. Thus, synthesis of genomes is sensitive to inhibition
templates. It is believed that synthesis of genomes from
by amanitin, requires S-HDAg that has been phosphorylated,
antigenomic templates and synthesis of the mRNA from
methylated, and acetylated, and the RNA product is imme-
genomic templates are carried out by RNA polymerase II.
diately exported to the cytoplasm. Synthesis may occur in
However, synthesis of antigenomes from genomic templates
the nucleoplasm. In contrast, synthesis of antigenomes is
may utilize another polymerase, perhaps RNA polymerase
resistant to amanitin, requires L-HDAg but does not require
I. Other, currently unknown host factors also participate,
its phosphorylation or acetylation, and the RNA product
and the HDAg, of which there are two kinds, S-HDAg and
is retained in the nucleus. Synthesis perhaps occurs in the
L-HDAg, as described later, both of which can be modified
nucleolus. Approximately 10 times as much genomic RNA
in various ways, is absolutely required. These replication
is produced as antigenomic RNA.
steps are illustrated schematically in Fig. 9.9A and B.
The genomic RNA is used as a template to produce the
Replication of the RNA, whether production of genomes
mRNA for the HDAg. The mechanism by which this RNA is
or antigenomes, is thought to utilize a rolling circle mech-
produced is not fully understood. It may resemble the process
anism in which concatenated RNAs are produced that are
for production of antigenomic RNA but occurs in a different
cleaved to unit length by the self-cleavage activity present
place in the nucleus utilizing a different RNA polymerase
in both genomic and antigenomic molecules. The resulting
and different forms of HDAg. Synthesis of mRNA is also
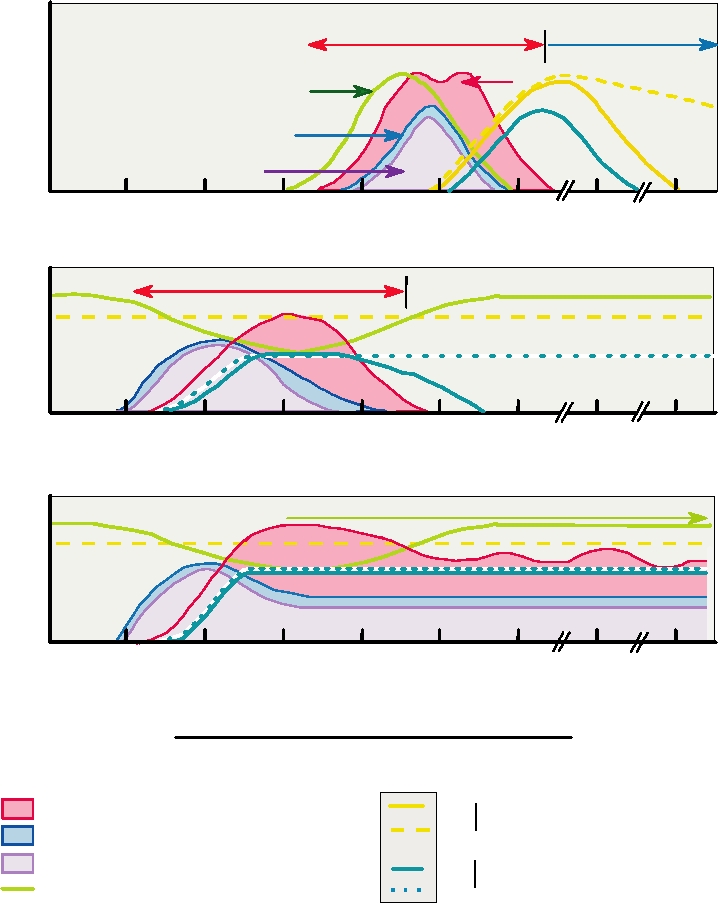
Simultaneous Coinfection with Hepatitis B and Hepatitis d
A.
Convalescence and
Acute Disease
Recovery
ALT
HBsAg
HDV RNA
HD Ag
0
2
4
6
8
10
12
24
32
Acute Hepatitis d after Superinfection of a Chronic Hepatitis B Patient
B.
Acute Disease
0
2
4
6
8
10
12
24
32
Chronic Hepatitis δ after Superinfection of a Chronic Hepatitis B Patient
C.
Severe chronic hepatitis B and chronic hepatitis δ
0
2
4
6
8
10
12
24
32
Weeks after exposure (coinfection or superinfection)
Serology
Antigens, RNA, and liver function markers
Antibody levels
IgM
ALT level (alanine aminotransferase)
Anti HBc (anti-hepatitis B core)
IgG
HD RNA (hepatitis d RNA)
HDAg (hepatitis d antigen)
IgM Anti HD (anti-hepatitis d antigen)
IgG
HBsAg (hepatitis B surface antigen)
FIGURE 9.7 Patterns of anti-hepatitis B core antigen antibody, hepatitis B antigen, hepatitis δ RNA, hepatitis δ antigen,
and ALT in patient serum during different types of coinfection with hepatitis δ and B. (A) Simultaneous infection by both
types. (B) and (C) Superinfection by hepatitis δ of a patient with chronic hepatitis B infection. Many infections start with
acute hepatitis δ as in (B). Some proportion of superinfections progress to chronic hepatitis with elevated liver enzymes
and sustained production of hepatitis δ RNA and protein as in (C). Adapted from Fields et al. (1996) p. 2825.
sensitive to amanitin, suggesting that RNA Pol II is involved
HDV Delta Antigen
and synthesis may occur in the nucleoplasm. There is a poly-
The mRNA for HDAg is exported to the cytoplasm and
adenylation site following the open reading frame (ORF)
translated into a polypeptide of 195 amino acids, referred to
for HDAg, and cellular enzymes are assumed to cut the pre-
as the small δ antigen or S-HDAg. This protein is required for
mRNA and polyadenylate it similar to what happens with
RNA replication. Thus, for example, in vitro systems to study
cellular mRNAs. The fact that the mRNA is also capped sug-
the replication of HDV RNA must be supplemented with
gests that the origin of synthesis may differ from that used for
S-HDAg for replication to occur. S-HDAg is a component of
RNA replication so that the process resembles cellular
the infecting particle and is therefore present in the infecting
production of mRNA rather than the replication of HDV RNA.
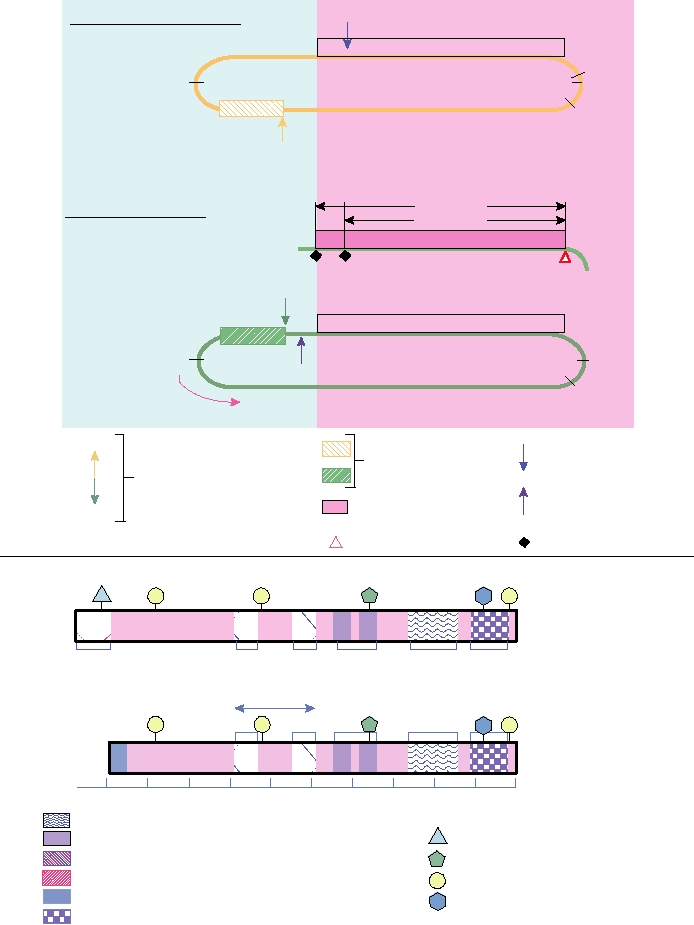
Viroid Domain
HDAg Coding Domain
1015
Genomic RNA ~1683 nt
(Oril)
1631
795
1638
0/1683
688/689
950 1017
1601
L-HD Ag
Antigenomic RNAs
S-HD Ag
5
3
0.8 kb mRNA
n
A
903/904
Antigenome
template
RNA
795
1638
An
0/1683
5
3
1015
688/689
RNA editing site
Ribozymes
Ribozyme cleavage sites
ORF for HD antigen
Polyadenylation site
903/904
An
Initiation codon
Termination codon
C211
S177
S123
K72
R13 S2
M
A
P
P
P
P
L-HDAg
(214 aa)
214195
146136 10797 8868
5231
272
RNA-binding
A
P
P
P
M
S-HDAg (195aa)
Amino acids
200
160
120
80
40
0
Posttranslational Modifications
Coiled-coiled sequence (dimerization signal)
Prenylation
Nuclear localization signal (NLS)
P
Arginine-rich motif (ARM)
Acetylation
A
Packaging signal
Phosphorylation
P
S-HD Ag-specific epitope
M
Methylation
Cryptic RNA-binding domain
FIGURE 9.8 Structures of HDV RNA and of the HDAg. Above horizontal line: schematic diagram of the structure
of HDV RNA. Nucleotides are numbered from the unique Hind III site in the cDNA clone of the prototype HDV.
Numbering is 5′ to 3′ in the genomic RNA. Nucleotides 795 and 1638 represent the ends of the rodlike structure. Below
line: schematic diagram of the structural and functional domains of the hepatitis delta antigen (HDAg). The protein is
shown in the same orientation as the mRNA in part (A), with the amino acids numbered from right to left. Other features
are described in the key. Adapted from Modahl and Lai (2000), and Lai (2005).
cell when RNA replication first begins. Production of new
thought to be effected by deamination, in the antigenome, of
protein after infection enables RNA replication to accelerate.
the adenosine in the UAG codon to produce inosine. A cel-
A second form of δ antigen is also produced during infec-
lular adenosine deaminase has been described that probably
tion. An RNA-editing event occurs in about one-third of
performs this function. Inosine pairs as guanosine, and con-
the antigenomic templates, in which the termination codon
tinued replication of the RNA will lead to the substitution of
UAG, at position 196 of the ORF for the δ antigen is changed
G for A. This RNA editing site is specific and requires spe-
to UGG, encoding tryptophan (see Fig. 9.8). This change is
cific sequences within the antigenomic RNA for it to occur.
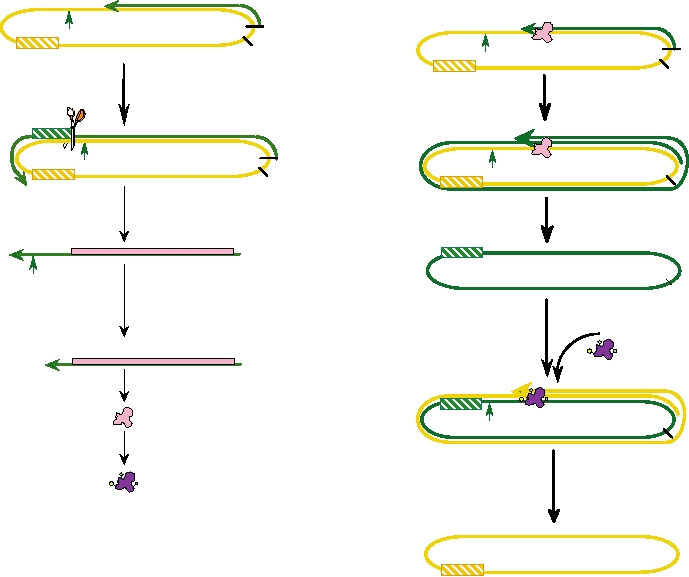
Replication of Hepatitis δ Genome RNA
B.
Transcription of Hepatitis δ mRNA
.
n the Nucleolus:
An enomic
G
RNA
5 1631
A
0/1683
Genomic RNA
0/168 5 1631
I
An
3
Transport to nucleoplasm
Pol I?
Pol II
Rolling circle?
03/904
HDAg
Genomic RNA
0/1683 5 1631
9
An
0/1683
An
Ribozyme cleavage
Ribozyme cleavage
Ligation
5
Processing and polyadenylation
Antigenomic RNA
An
0/1683
by host enzymes
Transport to nucleoplasm organelle
Transport to cytoplasm
Pol II
In the Nucleoplasm:
mHDAg
HDAg mRNA
T
Cap
An
ranslation
HDAg
Antigenomic RNA
An
Posttranslational modification
Rolling circle
mHDAg
Ribozyme cleavage
Ligation
Genomic RNA
FIGURE 9.9 Transcription and replication of hepatitis δ RNA. (A) Transcription of hepatitis δ mRNA. RNA synthesis
by Pol II begins about 30 nucleotides upstream of the HDAg ORF (at nt 1631). Plus-strand synthesis proceeds through
the ORF. The nascent strand is cleaved by the ribozyme at nt 903/904 and the mRNA is subsequently processed and
polyadenylated. HDAg mRNA is translated in the cytoplasm and some HDAg is posttranslationally modified (mHDAg).
(B) Replication of hepatitis δ genome RNA. Genome RNA is transported to the nucleolus where it is replicated by Pol I
in the presence of HDAg as a rolling circle. After ribozyme cleavage and ligation the antigenome RNA is transported out
of the nucleolus with mHDAg and genome-sense RNA is synthesized by Pol II in a second rolling circle, then cleaved
and ligated as before. Adapted from Lai (2005).
Change of the termination codon to a tryptophan codon
required for virus assembly, and isoprenylation is required
leads to the production of a polypeptide that is 19 residues
for this activity. A map of functional domains of the L- and
longer, for a total length of 214 amino acids, referred to as
S-HDAgs is shown in Fig. 9.8B.
the large d antigen or L-HDAg. Because editing is required,
it is only produced later in the infection cycle. The extent of
Assembly of Virus
editing is controlled, perhaps by S-HDAg. Obviously, only
genomes that are not edited can give rise to infectious viri-
Assembly of HDV virions begins with the formation of
ons. S-HDAg is required for replication, and only nonedited
a nucleocapsid or core that contains the HDV genome and
both the L and S forms of the δ antigen. The core is 19 nm
genomes encode it.
HDAg can be phosphorylated on Ser-2, Ser-177, and Ser-
in diameter and matures by budding, using the HBV sur-
123, methylated on Arg-13, and acetylated on Lys-72. These
face antigens. Budding appears to be the same as for HBV
modifications change the activities of the protein as well as
(Chapter 6), and the three surface antigens of HBV form the
its subcellular localization. As stated, S-HDAg is required
protein component of the outer envelope surrounding the
for RNA replication. L-HDAg can be isoprenylated on a
HDV capsid. Thus, although the RNA of HDV can replicate
cysteine four residues from the C terminus that is therefore
independently of HBV, assembly of progeny virions requires
not present in S-HDAg. L-HDAg suppresses RNA replica-
the simultaneous infection of the cell by HBV to supply
tion and leads to a shift from replication of RNA to encap-
the surface glycoproteins needed to produce infectious
sidation of RNA into progeny virions. It is specifically
particles.
Search WWH :