environment for reactivation of the virus rather than to result
highly diverse viruses that have been found in all mammals and
from infection by the virus. Nonetheless, these viruses are
birds that have been carefully examined, with the exception of
known to be capable of transforming cells and of causing
the laboratory mouse, and probably occur in most mammals
tumors in experimental animals, giving rise to the possibil-
and birds. The viruses are highly host specific. To date 118
ity, if not probability, that they cause tumors in humans.
virus types have been completely described, most of which,
Transformation of cells by JC virus appears to affect
for obvious reasons, are human viruses (HPVs). Cell culture
primarily cells of neuronal origin, and there is suggestive
systems for the study of these viruses are very limited because
evidence that in humans the virus may cause tumors in the
they undergo a complete replication cycle only in terminally
brain and perhaps in other organs as well. JC virus causes
differentiated cells, and most virus isolates are characterized
solid tumors in nonhuman primates as well as in rodents,
on the basis of nucleotide sequences of virus DNA obtained
giving further reason to believe it might be responsible for
directly from patients or infected animals. Only when the com-
some human cancer. For BK virus there also exists sugges-
plete genome has been characterized is the virus given a name,
tive evidence that the virus may cause a number of human
which consists of a sequential number together with a designa-
tumors.
tion for its host. Thus, although there are 96 recognized types
Early lots of poliovirus vaccine, both the inactivated Salk
of HPV, more exist and continue to be characterized.
vaccine and the live Sabin vaccine, were contaminated with
The classification of papillomaviruses has recently
live SV40 virus, which was a contaminant in the monkey
undergone major revisions. Classification is now based
kidneys used to produce the virus for the vaccines. A number
on the sequence of the most highly conserved protein in
of other live virus vaccines produced around this time were
the family, the major capsid protein called L1. If a virus
similarly contaminated with SV40. As a result, many mil-
that differs by more than 10% in the nucleotide sequence
lions of people were infected with the virus. SV40 shares
encoding this protein from that of the virus to which it is
69% sequence identity with JC and BK viruses (Table 7.13),
most closely related, it is recognized as a different type and
and causes tumors in experimental animals, giving rise to
given a new number. Isolates that differ by 2 to 10% in the
concern that it might cause tumors in humans. Extensive
sequence of this gene are considered to be different subtypes
study of the cohort of people infected as a result of contami-
of the same virus type, and if they differ by less than 2%
nated vaccines, both in the United States and in Europe, has
they are classified as different strains of the same virus type
not revealed any convincing evidence for tumors associated
or subtype. Grouping into genera and species is also based
with SV40 infection, although suggestive evidence exists
on the sequence of the L1 gene. Viruses that share less than
that implicates SV40 in a number of tumors, especially brain
60% nucleotide sequence identity in the L1 gene are classi-
tumors and mesothelioma tumors of the lungs. The prob-
fied into different genera, and species within a genus share
lem is complicated by the fact that SV40 now circulates in
between 60 and 70% sequence identity. Thus, virus types
the human population, whether as a result of its introduc-
within a species share 7190% identity. This definition of a
tion with the poliovirus vaccine or from another source is
species is biologically relevant, because it groups virus types
not known. It is also complicated by the fact that although
that share important biological traits.
the virus is often found associated with certain tumors, it is
Sixteen currently recognized genera of papillomaviruses
not found in all such tumors. Is the association with such
are listed in Table 7.14. Genera are simply named by Greek
tumors, therefore, adventitious or causative? The case of
letters. The HPVs fall into 5 genera at present, only one of
mesothelioma illustrates this point. These tumors are clearly
which, Alphapapillomavirus, is known to contain a nonhu-
associated with exposure to asbestos but SV40 is often asso-
man virus as well as human viruses, but even in this case all
ciated with them. Is the virus a cofactor in the development
of the known viruses in this genus are primate viruses. The
of the tumor or simply a freeloader? It should be noted that
96 HPV types are grouped into 27 species using the rules out-
in animal models polyomaviruses are most likely to cause
lined earlier. Each species is named after the prototype HPV
tumors in animals that are nonpermissive for virus replica-
type in that species. Thus, for example, 14 species are recog-
tion. SV40 is primarily a monkey virus and human cells are
nized in the genus Alphapapillomavirus, most of which con-
only semipermissive for virus replication, suggesting that
sist of multiple virus types as defined before. As an example
this virus may be more likely to cause tumors in humans
of one of the species within the genus, Alphapapillomavirus
than the human viruses BK and JC.
species 16, this species contains HPV types 16, 31, 33, 35,
52, 58, and 67. Beta-, Gamma-, Mu-, and Nupapillomavirus
genera contain 5, 5, 2, and 1 species, respectively.
FAMILY PAPILLOMAVIRIDAE
Infection by Papillomaviruses
Papillomaviruses resemble polyomaviruses in structure but
are larger (Fig. 2.5). The virion is 55 nm in diameter, and the
Papillomaviruses infect epithelial cells, either mucosal
circular dsDNA genome is 8 kb in size. Papillomaviruses are
or cutaneous. Each virus is usually more or less restricted
TABLE 7.14 Papillomaviridae
Virus name
Usual
Genus/members
abbreviation
host(s)
Disease/genome characteristics
Alphapapillomavirusa
Human papillomaviruses 2, 6, 7, 10, 16, 18, 26,
HPV-2, etc.
Humans
Oral and anogenital mucosal lesions
32, 34, 53, 54, 61, 71, cand 90
E5 ORF conserved between early and late regions
Rhesus monkey papillomavirus 1
RhPV-1
Primates
Betapapillomavirus
Human papillomaviruses 5, 9, 49, cand 92, cand 96
HPV-5 etc.
Humans
Epidermodysplasia verruciformis
activated by immunosuppression
Gammapapillomavirus
Human papillomaviruses 4, 48, 50, 60, 88
HPV-4 etc.
Humans
Cutaneous lesions
E5 ORF is absent
Deltapapillomavirus
European elk papillomavirus
EEPV
Elk
Fibropapillomas
Other ovine and bovine papillomaviruses
Cattle, sheep and deer
Epsilonpapillomavirus
Bovine papillomavirus 5
BPV-5
Cattle
Cutaneous papillomas
Zetapapillomavirus
Equine papillomavirus 1
EcPV
Horses
Cutaneous papillomas
Etapapillomavirus
Chaffinch papillomavirus
FcPV
Birds
Cutaneous lesions
E6 ORF absent
Thetapapillomavirus
Timneh African gray parrot papillomavirus
PePV
Birds
Cutaneous lesions
E4, E5, E6 ORFs are absent
Iotapapillomavirus
Mastomys natalensis papillomavirus
MnPV
African soft-furred rat
Cutaneous lesions
E2 ORF larger and E5 ORF absent
Kappapapillomavirus
Cottontail rabbit papillomavirus
CRPV
Cottontail rabbits
Cutaneous and mucosal lestions
E6 larger, and extra E8 ORF
Lambdapapillomavirus
Canine oral papillomavirus
COPV
Dogs
Cutaneous and mucosal lesions
Feline papillomavirus
FdPV
Cats
Region between early and late genes very long
Mupapillomavirus
Human papillomaviruses 1, 63
HPV-6, -63
Humans
Cutaneous lesions
Nupapillomavirus
Human papillomavirus 41
HPV-41
Humans
Benign and malignant cutaneous lesions
Xipapillomavirus
Bovine papillomavirus 3
BPV-3
Cattle
True papillomas
ORF 6 absent
Omikronpapillomavirus
Phocoena spinipinnis papillomavirus
PsPV
Cetaceans
Genital warts
ORF 7 absent
Pipapillomavirus
Hamster oral papillomavirus
HaOPV
Hamsters
Mucosal lesions
a
Transmission in most cases is by close contact, including sexual contact and the viruses are found worldwide. For human viruses, viruses with red numbers
cause malignancies, while those in blue are primarily benign. Nonhuman virus names are colorcoded according to their hosts.
to specific sites in the host. The receptors for the virus are
Transcription of mRNAs
unknown, but they enter the cell by receptor-mediated endo-
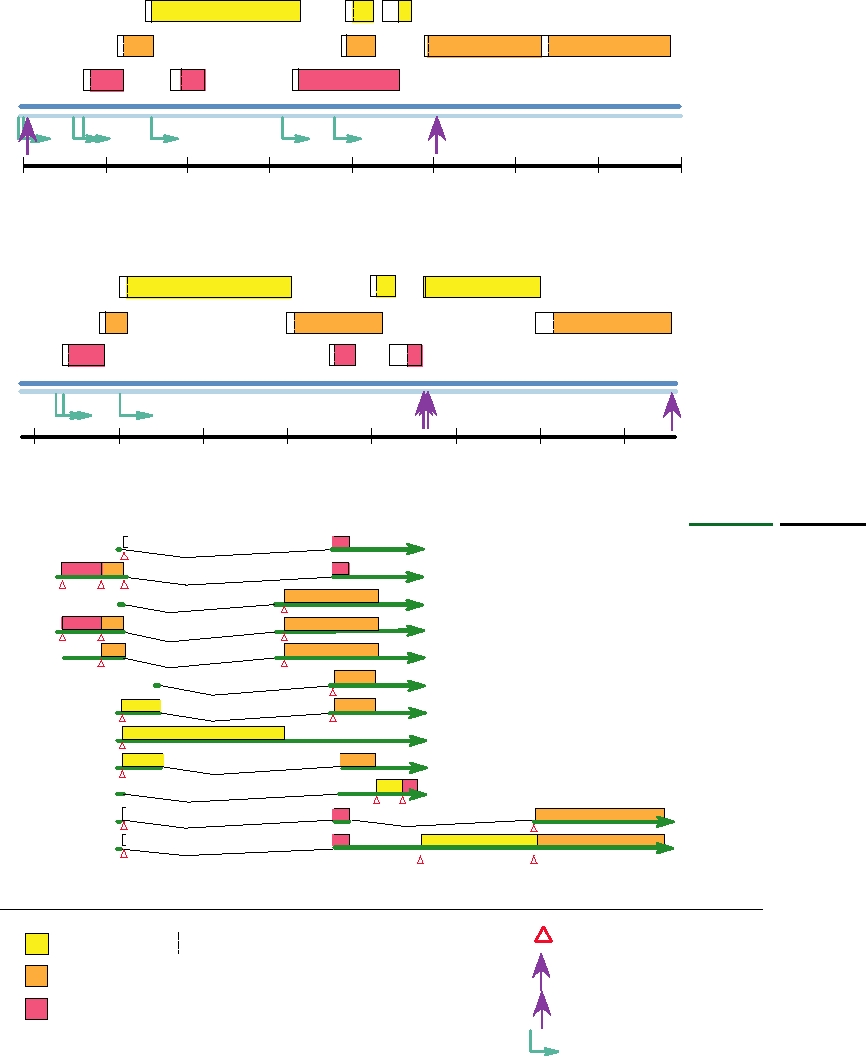
The genome organizations of two papillomaviruses, BPV-
cytosis. In order to enter the cytoplasm from the endosomal
1 and HPV-11, are shown in Fig. 7.30A and B and a detailed
compartment, cleavage of the viral minor capsid protein L2
transcription map of HPV-11 showing the exons, transcrip-
by furin is required. This is the same protease that cleaves
tional promoters, and polyadenylation sites is shown below
the glycoproteins of many enveloped viruses to render them
in Fig. 7.30C. Unlike the polyomaviruses, all papillomavi-
infectious, but here it is used by the virus upon entry rather
rus mRNAs are transcribed in the same direction from only
than during assembly of the virion. Although furin is a type
one of the two strands. The genomes of papillomaviruses
I membrane protein that is known to be present in the trans-
are circular, but the genomes have been linearized in the
Golgi network, it is also present on the cell surface, and it is
figure for ease of presentation. In the absence of cell culture
probably here that it cleaves L2.
systems for the virus, most studies of RNA transcription
The virus first infects cells of the basal proliferative
have used RNA extracted from papillomas or from carci-
layer of epithelium, probably at the site of a cut or abrasion
noma cell lines. Because of this, the maps are probably not
that gives it access to this layer. The viral DNA enters the
complete.
nucleus where it is maintained as a low-copy number plas-
The transcription of papillomavirus RNAs is complex, as
mid and only the early genes are expressed. When the cells
illustrated in the figure. There are multiple promoters and splice
divide, the viral DNA is transmitted to both daughter cells
sites and differential use of these in different cells. Furthermore,
by means of tethering the DNA to mitotic chromosomes
there is extensive overlap of genes in the genome. Different
by the E2 protein. This mechanism of insuring distribu-
reading frames encoding different peptide sequences may be
tion of viral DNA to progeny cells is also used by two her-
linked in different ways by alternative splicing.
pesviruses, EBV and HHV8. Only when the cells become
In BPV-1 there are at least seven promoters for transcrip-
terminally differentiated does the amplification of the viral
tion of RNA and more than 20 different mRNAs have been
DNA begin in earnest, the expression of late genes com-
identified. Six promoters are used for the transcription of
mences, and the progeny virions are assembled and shed.
early mRNAs, all of which terminate at a poly(A) addition
Our knowledge of the replication of the papillomavi-
site at position 4180 (AE). The seventh promoter is used for
ruses is limited because none will undergo a full replication
the transcription of the late mRNAs, which terminate at a
cycle in any simple tissue culture system. Bovine papil-
poly(A) addition site at 7156 (AL). The early genes of BPV-
lomavirus (BPV-1) has been the most extensively charac-
1 include E1 and E2, required for DNA replication, and
terized because it proliferates in dermal cells as well as in
E5, E6, and E7, required for cell transformation. The late
terminal epithelial cells. It readily infects and transforms
genes, which are expressed only in terminally differentiated
rodent cells, in which the early proteins are expressed and
epithelial cells, include L1 and L2, which encode proteins
viral DNA replication occurs. In transformed cells, the
present in the virion. L1 is the major capsid protein and
BPV genome is maintained as a stable plasmid and this
when expressed alone assembles into virus-like particles
feature has permitted the use of BPV-1 as an expression
that appear identical to virions. If L2 is coexpressed with L1,
vector. Much effort has also been put into the study of the
it is also incorporated into the virus-like particles. Another
human papillomaviruses (HPVs) because of their associa-
late gene is E4, which although located in the early region is
tion with human cancer, but these studies have been ham-
expressed from the late promoter.
pered because human papillomaviruses will only grow in
The pattern of transcription in HPV-11, illustrated in Fig.
humans and human cells and will only undergo a full lytic
7.30C, is slightly different. Only three promoters are known,
cycle in terminally differentiated cells. Two recent devel-
two for the early genes and one for the late genes. There are
opments have helped in the study of HPVs. A tissue culture
poly(A) addition sites for early and late transcripts. Proteins
systems has been developed in which epidermal cells will
corresponding to those of BPV-1 are produced during HPV-
differentiate, permitting at least limited studies of a full
11 infection, but the complexity of the pattern of proteins
growth cycle. This method is laborious, however. Another
produced and the difficulties in studying HPV replication
approach has been the development of packaging sys-
make exact comparisons difficult. However, three regions
tems in which viral DNA and proteins are expressed from
of the genome are recognized, the early region encoding
expression plasmids, and large numbers of virus particles
nonstructural proteins (about 4 kb of DNA), the late region
are formed. Other studies have simply used expression of
encoding the structural proteins (about 3 kb of DNA), and
various viral proteins from expression vectors to study the
the noncoding long control region or upstream regulatory
properties of these proteins, or have used grafts of infected
region of about 1 kb that contains cis-acting elements that
human tissue in immunocompromised mice to study a full
regulate viral replication and gene expression.
replication cycle.
A.
Bovine Papillomavirus 1 (7945 bp)
E1
E3
E5
E4
L2
L1
E7
E6
E8
E2
LCR
DNA
L
E
7945/0
1000
2000
3000
4000
5000
6000
7000
B.
Human Papillomavirus 11 (7933 bp)
E5a
L2
E1
E2
E7
L1
E6
E4
E5b
URR
DNA
L
E
7933/0
1000
2000
3000
4000
5000
6000
7000
C.
Transcription Map of HPV 11
Proteins
mRNAs
a
E1i^E4
CAP
An
CAP
b
E6, E7, E1i^E4
An
CAP
c
E2
An
CAP
d
E6, E7, E2
An
CAP
e
E7, E2
An
CAP
f
E2c
An
CAP
g
E1M, E2C
An
CAP
h
E1
An
CAP
i
E1M^E2C
An
CAP
j
E5a, E5b
An
CAP
k
E1i^E4, L1
An
CAP
l
E1i^E4, L2, L1
An
Translation initation
Most 5 AUG codon in an ORF
ORF 1
L Late polyadenylation site
LCR Long control region
ORF 2
URR Upstream regulatory region
ORF 3
E Early polyadenylation site
Transcriptional promoter
FIGURE 7.30 Genome organization and transcription map of papillomaviruses. (A) Genome organization of bovine
papillomavirus 1. (B) Genome organization of human papillomavirus 11. (C) Transcription map of human papillomavirus
11. Genomes have been linearized at the upstream regulatory region (LCR or URR) for ease of presentation. All ORFs are
transcribed from left to right from one DNA strand. Promoters are shown as turquoise arrows, sites of poly(A) addition with
purple arrows labeled late (L) or early (E), initiation codons are shown as open red triangles. The symbol "∧" joins two
ORFs that are translated together as a fusion protein from a spliced RNA. Adapted from Nathanson et al. (1996), p. 27 and
from Fields et al. (1996), pp. 2051 and 2052.
have a short N-terminal cytoplasmic domain, a transmem-
DNA Replication
brane region, and a more extensive C-terminal extracellular
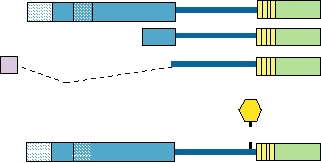
DNA replication requires the activities of E1 and E2.
domain. It activates the receptor for platelet-derived growth
The E1 protein binds to the origin of replication, has heli-
factor, perhaps by binding to it and causing it to dimerize.
case activity, binds α-primase, and is presumed to promote
Dimerization of many growth factor receptors present at the
the initiation of DNA replication. Thus, it has many of the
surface of cells results in activation of a protein kinase and
functions of the polyomavirus T antigens. E2 is a regulatory
phosphorylation of tyrosines, which leads to the activation of
protein that is produced in multiple forms (Fig. 7.31). It can
transcription factors whose activities stimulate cell prolifera-
either transactivate or repress genes, depending on the loca-
tion. BPV-1 E5 also binds other cellular proteins that may
tion of binding sites for it within a gene. It plays an important
be involved in transformation. Transformation of established
role in the regulation of transcription, and interacts with E1
rodent cells as defined by a number of criteria can be obtained
to promote recognition of the promoter for efficient replica-
by expression of E5 alone, but the expression of both E6 and
tion of DNA. The interaction with E1 may help recruit host
" is required for the fully transformed phenotype.
replication factors to the origin of replication or promote the
The E5 encoded by HPVs has also been shown to induce
assembly of the preinitiation complex.
some transforming alterations, but whether this protein plays
DNA replication occurs in two phases. In cells that the
an important role in transformation is uncertain. More is
virus infects nonproductively (cells transformed by BPV-1
known about HPV E6 and E7. Expression of E6 and E7 from
or cells in the dermal layer of the epithelium), the DNA is
high-risk strains of HPV (strains that are often associated
maintained as a multiple copy plasmid (50400 copies per
with human cancer) are capable of facilitating the immor-
cell). After replicating sufficiently to reach this number of
talization of primary human keratinocytes. E7 is a small
copies, further DNA replication is limited to that required
zinc-binding protein that is phosphorylated and is capable
to maintain the copy number as cells divide. A complete
of transforming cells when expressed alone. It binds the
replication cycle occurs only in terminally differentiated
cellular tumor suppressor protein Rb as well as p107 and
cells, where large numbers of DNA genomes are produced
p130 (Fig. 7.22). Rb undergoes changes in phosphorylation
for incorporation into progeny virions. Since these termi-
induced by cyclin-dependent kinases at the GS1 boundary.
nally differentiated cells do not divide, they are intrinsically
In its hypophosphorylated form it inhibits cell cycle progres-
incompetent in supporting DNA synthesis. Thus, large-scale
sion. The viral oncogene preferentially binds the hypophos-
production of viral DNA genomes in these cells requires the
phorylated form, thus preventing its inhibitory activity and
activity of viral transforming genes.
inducing cycling of the cell (and therefore DNA synthesis).
Genetic studies have shown that binding to Rb is required
in order for E7 to transform cells. It is of considerable inter-
The Transfor ming Genes
est that E7 from low-risk HPVs binds Rb only one-tenth as
Three genes of papillomaviruses, E5, E6, and E7, have
efficiently as E7 from high-risk strains, and that the E7 from
been shown to be involved in transforming cells. E5 of BPV-
low-risk strains is inefficient in transformation assays.
1 is a small polypeptide (44 amino acids) that is believed to
E6 from high-risk HPVs, but not from low-risk HPVs,
can complex with the tumor suppressor protein p53 (Fig.
7.22). SV40 and adenovirus oncoproteins also bind p53, but
Transactivation
Hinge DNA-Binding
simply sequester it. In contrast, binding of HPV E6 leads
Domain
Domain
to the degradation of p53 by the ubiquitin-mediated degra-
dation pathway. Removal of p53 has the effect of inducing
Protein E2 (48KD)
1
DNA synthesis, as described earlier. The importance of Rb
Protein E2-TR (31KD)
and p53 in regulating cell cycling is made clear by the fact
162
E8
Protein E8^E2 (28KD)
206
that three different viruses--high-risk HPVs, adenoviruses,
and SV40--all target these proteins in order to provide an
P
atmosphere conducive for DNA replication by the virus.
S Basic region
HPV E6 can also activate transcription of hTERT, the cata-
Hydrophobic
Amphipathic
lytic subunit of telomerase. Full transformation of human
repeats
helices
epithelial cells requires telomerase expression as well as the
expression of viral and cellular oncogenes.
FIGURE 7.31
Structure of BPV-1 virus E2 protein. Full-length E2
contains a transactivation domain at the N terminus, linked by a hinge to
a DNA-binding domain at the C terminus. There are 3 forms of E2, all of
Papillomaviral Disease
which contain the 85amino acid long DNA-binding domain. The bottom
diagram identifies other functional features of E2 such as the amphipathic
On infection, papillomaviruses induce cellular prolif-
helices, hydrophobic repeats, a basic region, and a phosphorylation site.
eration that leads to the production of warts or papillomas.
Data from McBride et al. (1989).
The viruses are usually restricted to a specific area of the
respiratory tract. The warts are normally self-limited
body such as the skin, the mouth, the throat, or the geni-
proliferative lesions that regress after some time because of
tal tract. In most infections these eventually resolve, but in
an immune response. Cytotoxic T lymphocytes are thought
some cases tumors can result. Papillomaviruses of humans,
to play an important role in regression, and warts are often
cattle, sheep, and cottontail rabbit have been shown to be
more numerous under conditions where the immune system
associated with cancers in their natural hosts.
is suppressed. HPVs are spread by direct contact and infection
begins at the site of an abrasion in which the virus can contact
the deeper epithelial layers.
HPVs are not only specific for humans but also for the
Human Papillomaviruses
tissues infected. HPVs cause either cutaneous lesions or
More than 200 different strains of HPV have been identified
mucosal lesions. Eleven of the HPV species shown in Table
by analyses of full or partial sequences of viral DNAs isolated
7.15 cause skin warts. Only a few of these are responsible
from individual lesions and classified into 96 different types
for most skin warts, which are common in school children.
to date. These 96 types, numbered from 1 to 96, are classi-
The remainder have been isolated only from patients who
fied in turn into 27 species found in 5 different genera. Species
suffer from a rare disease called epidermodysplasia verru-
are named after the type within that species with the lowest
ciformis. These patients are unable to resolve their warts,
number, which is therefore the first described type within that
probably because of an inherited immunologic defect, and
species, and species numbering is thus not sequential. A list-
wartlike lesions appear all over the body. These warts often
ing of 19 of these species that shows the tissues infected and
become malignant after many years, especially on areas of
the probability that infection leads to cancer is shown in Table
the skin exposed to sunlight, but these tumors are generally
7.15. HPVs cause warts in the skin, genital tract, mouth, or
slow growing and do not metastasize.
TABLE 7.15
Common Clinical Lesions Associated with Human Papillomaviruses
Disease
Type/isolates
Anatomical site
Common name
Medical term
Risk
Cancers
Cutaneous HPVs
HPV 1, 4
Sole, palm
Plantar warts
Verruca plantaris
None
None
HPV 2, 4, 10, 26
Cutaneous
Common warts
Verruca vulgaris
None
None
HPV 2, 5, 10
Cutaneous
Flat warts
Verruca plana
None
None
Skin carcinomasa
HPV 5, 50
Face, trunk, esophagus
Benign warts
EV
High
HPV 5, 9
Cutaneous
Flat warts
Verruca plana
In ISP
HPV 5, 9
Cutaneous
Macular lesions
EV (benign)
Some
In ISP
HPV 5, 9, 49
Cutaneous
EV
Some
Some SCC
Skin carcinomasa
HPV 9
Cutaneous
Malignant melanoma
High
HPV 41, 48
Cutaneous
SCC
Some
In ISP
HPV 49
Cutaneous
Plantar warts
Verruca plantaris
?
In ISP
Mucosal-Associated HPVs
Rareb
HPV 6
Anogenital, larynx
Genital warts
CD
Low
Rareb
HPV 6, 7, 32, 34
Anogenital
Anogenital warts
IN
Low
HPV 6, 7, 26, 61
Genital
Genital warts
CIN
Low
HPV 61, 34
Larynx
Oral papillomas
Low
In ISP
HPV 26
Genital mucosa
Genital warts
CIN
Some
Some malignant progression
HPV 16, 18, 53
Genital mucosa
Genital warts
CIN
High
13% progress to cervical
carcinomas, cofactors unknown
a
3040% undergo neoplastic conversion in sun-exposed area.
b
On rare occasions these HPV types have also been found associated with carcinoma.
Abbreviations: SCC, squamous cell carcinoma; EV, epidermodysplasia verruciformis; CD, condyloma acuminatum; CIN, cervical intraepithelial neoplasia;
IN, intraepithelial neoplasia; ISP, immunosuppressed patients.
Source: Adapted from Fields et al. (1996), Table 1, pp. 20482049, Tables 3 and 4; p. 2085; and Alani and Münger (1998).
Search WWH :