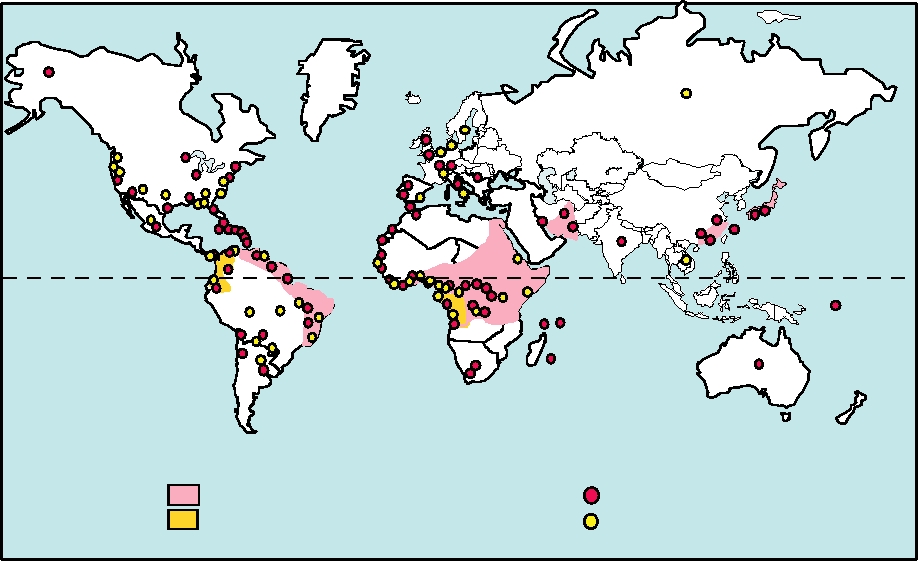
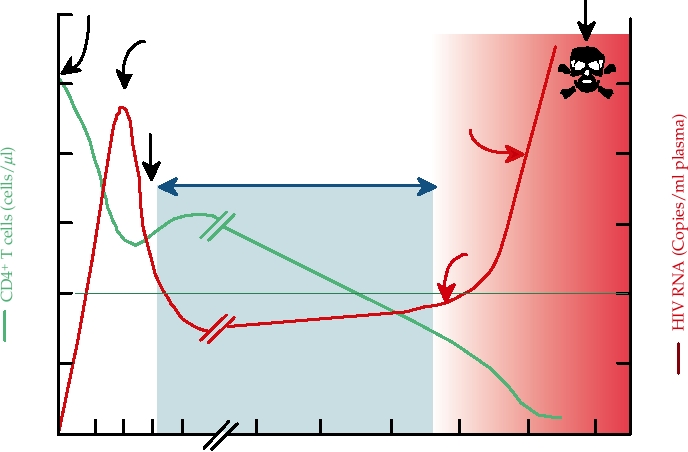
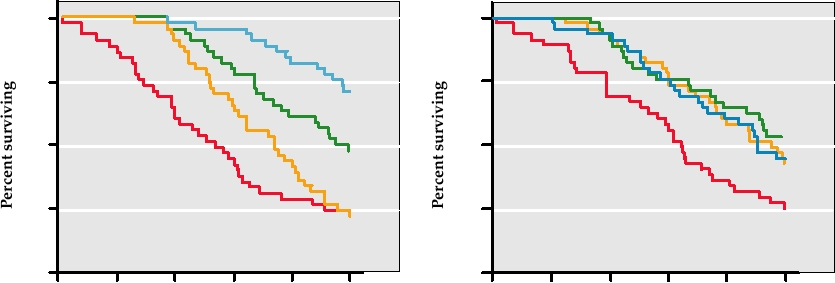
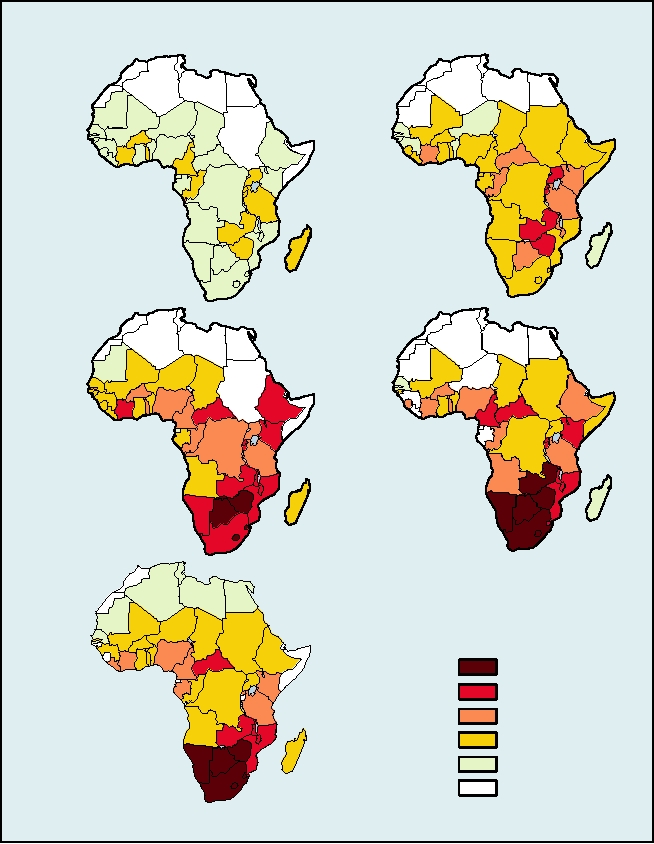
The most important hepadnavirus is hepatitis B virus,
nation with the host or with other viruses. Based on these
which is a major cause of hepatitis in humans. Like hepatitis
sequence relationships, the retroviruses that infect birds and
C virus, it often establishes a chronic infection that can result
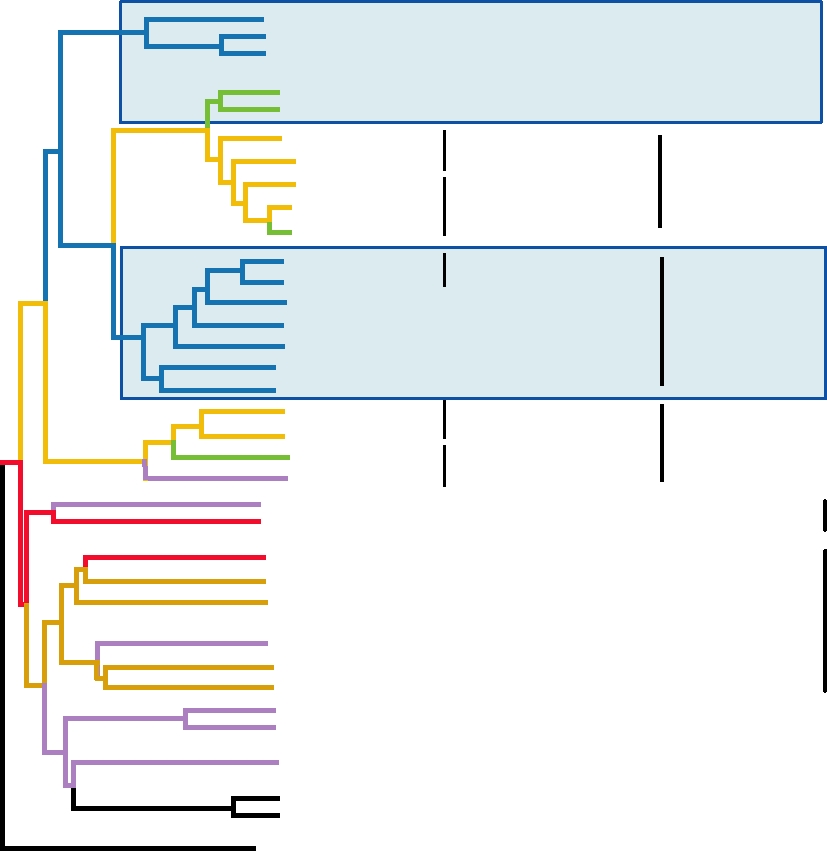
mammals are classified into six genera, as illustrated in Fig.
in cirrhosis or hepatocellular carcinoma.
6.1 and as listed in Table 6.1. Of these, members of three gen-
era are characterized as simple retroviruses, which encode
only the genes gag, pro, pol, and env (and sometimes dut).
FAMILY RETROVIRIDAE
The other three genera of retroviruses of higher vertebrates, as
well as the fish viruses, encode, in addition, regulatory genes
The retroviruses are a very large group of viruses that
that control their life cycle, and they are called complex ret-
infect invertebrates as well as vertebrates. Most of what
roviruses. Notice that these regulatory genes independently
we know about this group of viruses comes from studies of
entered the four different lineages of complex retroviruses
viruses that infect birds or mammals. Hundreds have been
represented by the four different genera (Fig. 6.1). Thus,
studied and, although considerable divergences exist, they
recombination to acquire new functions has been an ongo-
form a well-defined taxon. All are sufficiently similar to be
ing process in the retroviruses. Also notice that the complex
classified as belonging to a single family, the Retroviridae.
retroviruses do not group together. The epsilonretroviruses
The family gets its name from the concept that these viruses
are more closely related to the gammaretroviruses, which are
use retrograde flow of information, from RNA to DNA,
simple viruses, than they are to other complex retroviruses,
whereas the conventional flow of information in living
and the deltaretroviruses, lentiviruses, and spumaviruses are
organisms is from DNA to RNA.
not particularly closely related.
The RTs of retroviruses are the most highly conserved
Members of the different genera differ in their structure
elements of these viruses and have been used to study the
as visualized in the electron microscope. The simple viruses
relationships among them. Figure 6.1 illustrates the relation-
were formerly classified on the basis of morphology into
ships among the retroviruses of higher vertebrates based on
groups A, B, C, and D. The nucleocapsids of C-type viruses,
the sequences of their RTs. Included in the tree is a line-
now classified as alpharetroviruses and gammaretroviruses,
age of fish viruses, now classified as members of the genus
assemble during budding, and the nucleocapsid is centrally
Epsilonretrovirus. The tree is annotated to show where vari-
located in the mature virion. The nucleocapsids of B-type
ous new genes entered different virus lineages via recombi-
and D-type viruses, now classified as betaretroviruses,
Virus
Genus
MLV
new env
FeLV
Gammaretrovirus
HERV-C
WDSV
Epsilonretrovirus
orfA, orfB, orfC
HSRV
Spumavirus
bel1, bel2
HIV-1
tat, rev
HIV-2
Lentivirus
EIAV
VMV
dut
MPMV
new env
sag
Betaretrovirus
MMTV
HERV-K
IAP
RSV
Alpharetrovirus
BLV
PTLV-1
Deltaretrovirus
tax, rex
PTLV-2
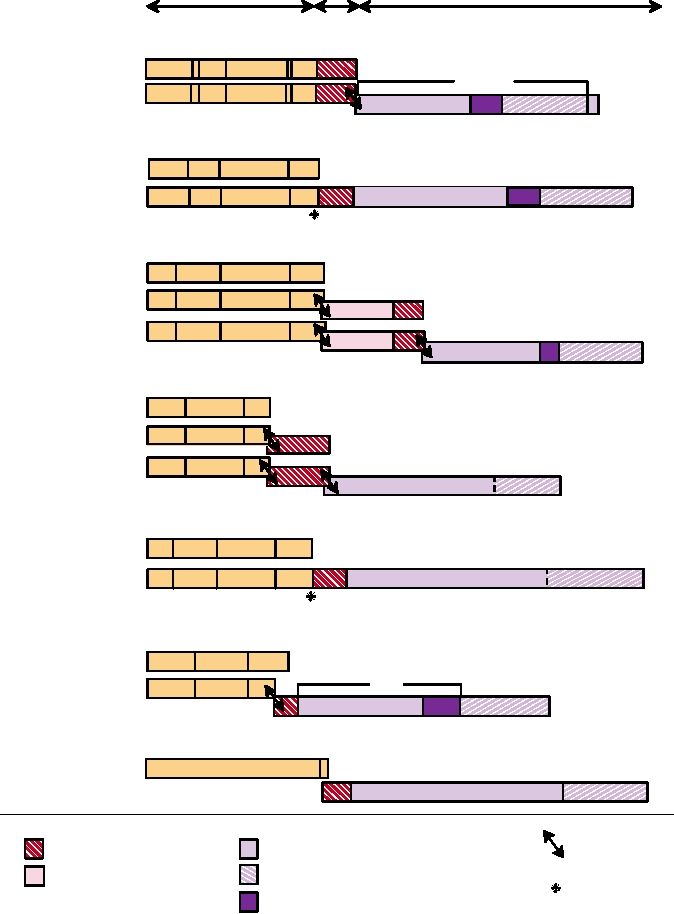
FIGURE 6.1
Phylogenetic tree of the Retroviridae drawn from the amino acid sequences of the reverse transcriptases.
The lengths of the branches are proportional to the degree of divergence; the names of the "simple" retrovirus genera
are boxed in black, the "complex" genera are boxed in red. The greenish ovals indicate the acquisition of new genes
during the evolution of current extant viruses. Most of the virus name abbreviations are found in Table 6.1; HERV-C and
HERV-K are defective retroviruses in the human genome and IAP is a virus-like element in rodent genomes. Adapted
from Coffin et al. (1997) Figure 6 on p. 43. and Fields et al. (1996) p. 1769.
TABLE 6.1 Retroviridae
Virus name
World
Genus/members
abbreviation
Host(s)
Transmission
Disease
distribution
Orthoretrovirinae
Alpharetrovirus (simple) formerly avian type C retroviruses
Avian leukosis
ALV
Birds
Worldwide
Rous sarcoma
RSV
Birds
Gammaretrovirus (simple) formerly mammalian type C retroviruses
Moloney murine leukemia
MLV
Mice
T-cell lymphoma
Feline leukemia
FeLV
Cats
T-cell lymphoma,
immunodeficiency
Betaretrovirus (simple) formerly mammalian type B and type D retroviruses
Mouse mammary tumor
MMTV
Mice
Vertical, including
Mammary carcinoma,
Worldwide
mothers' milk
T-cell lymphoma
Mason-Pfizer monkey
MPMV
Monkeys
Unknown
Deltaretrovirus (complex) formerly the BLV/HTLV group
Bovine leukemia
BLV
Cows
B-cell lymphoma
Primate T-lymphotropica
PTLV-1
Humans
Vertical, including
T-cell lymphoma,
mothers' milk, sexual
neurological
transmission, blood
disorders
TSP, HAMb
PTLV-2
Humans
Epsilonretrovirus (complex)
Walleye dermal sarcoma
WDSV
Fish
Benign sarcomas
North America
Lentivirus (complex)
Human immunodeficiency
HIV-1
Humans
Neonatal infection,
AIDS
Worldwide
HIV-2
Humans
sexual transmission, blood
Simian immunodeficiency
SIV
Monkeys
Simian AIDS
Africa
Visna-maedi
VISNA
Sheep
Neurological disease
No. Europe
Equine infectious anemia
EIAV
Horses
Anemia
Current epidemic in
Utah
Spumaretrovirinae
Spumavirus (complex)
Chimpanzee foamy
CFV
Monkeys
?
None
Human spumaretrovirus
HRSV
Humans
?
a
Primate T-lymphotropic virus 1 (PTLV-1) was formerly known as T-cell leukemia virus (HTLV).
b
TSP, tropical spastic pareparesis; HAM, HTLV-associated myelopathy.
assemble before budding and the nucleocapsid is eccentri-
evolving with the eukaryotes for a long period of time. The
cally located (type B) or bar shaped (type D) in the mature
integrated copies of retroviruses in the germ line constitute
virion (see Figs. 2.1 and 2.21).
a form of fossil record that allows us to trace the lineage of
Eukaryotic genomes contain a very large number of
at least some retroviruses for 100 million years or longer.
genetic elements that are related to retroviral genomes.
For other viruses, whether RNA or DNA, we can only trace
Retrotransposons encode RT and can move around within
ancestry for much shorter periods of time.
the genome by a process that uses reverse transcription and
insertion, similar to what happens with retroviruses. They
Retroviral Genome
are related to retroviruses but have no independent lives as
viruses. Other elements in the eukaryotic genome contain
The RNA genome in the retrovirion is diploid, consisting
additional retrovirus-like genes and appear to have arisen by
of two copies of a 7- to 10-kb single-stranded (ss)RNA that
insertion of retroviral genomes into the germ line at some
is capped and polyadenylated. The two copies of the genome
time in the past. Some of these are still active, capable of
are normally identical, but during mixed infection hybrid
genomes can result. They are joined near their 5˘ ends, and
giving rise to infectious retroviruses, whereas others are
defective. It is clear that this class of elements has been co-
perhaps in other regions as well, by hydrogen bonds.
Reverse Transcription of Viral RNA
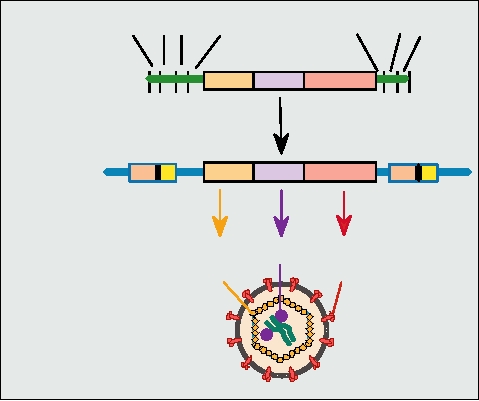
All retroviruses encode the four genes called gag, pro, pol,
and env, which are always found in this order in the genome.
A schematic overview of the life cycle of a retrovirus
A simplified illustration of a retroviral genome, its relation to
was presented in Fig. 1.13. Most retroviruses penetrate into
the provirus, and the locations of the different gene products
a cell by fusion with the plasma membrane, but some use the
in the virion are shown in Fig. 6.2. The name gag comes from
endosomal pathway. Once inside the cell, reverse transcrip-
group-specific antigen, because the proteins encoded in this
tion takes place in a subviral particle to produce a full-length,
gene are more highly conserved, and therefore more widely
linear dsDNA copy of the RNA. The composition of this sub-
cross-reactive immunologically, than are the envelope pro-
viral particle is not well defined. It certainly contains RT and
teins of the virion. The peptides derived from the Gag poly-
its associated activities as well as components derived from
protein, the precursor polyprotein encoded in the gag gene,
Gag, but which Gag components are present and whether
form the capsid of the retrovirion. The pro gene encodes a
cellular proteins form part of this particle are not known.
protease (PR) that is required for the processing of Gag, and
Reverse transcription begins in the cytoplasm. In most
pol encodes three activities, RT, RNase H, and integrase (IN).
viruses, the full-length dsDNA is produced in the cytoplasm,
RNase H forms a separate domain in the RT-RNase H protein,
but in some the finishing touches occur after transfer to the
but functions as an integral component of RT. IN is required
nucleus. After transfer to the nucleus, the viral DNA is inte-
for the integration of the dsDNA copy of the retroviral genome
grated into the host chromosome, essentially at random, in a
into the host chromosome. The fourth gene, env, encodes the
process that requires the activity of IN. In the simple viruses,
envelope glycoproteins present at the surface of the enveloped
transfer to the nucleus occurs during cell division, when
retrovirion. The primary product is Env, which is processed
the nuclear envelope is disassembled. At least some of the
by cleavage to form an N-terminal external protein called SU
complex retroviruses, however, such as HIV, encode pro-
(for surface) and a C-terminal protein that spans the mem-
teins that allow the DNA-containing complex to traverse the
brane called TM (for transmembrane). The order of genes in
nuclear membrane. Thus, the simple retroviruses can only
the provirus is the same as in the viral genome.
productively infect dividing cells, whereas HIV can infect
As described earlier, the genomes of complex retroviruses
nondividing cells.
contain a number of regulatory genes in addition to these
four basic genes present in all retroviruses. These genes will
be described later.
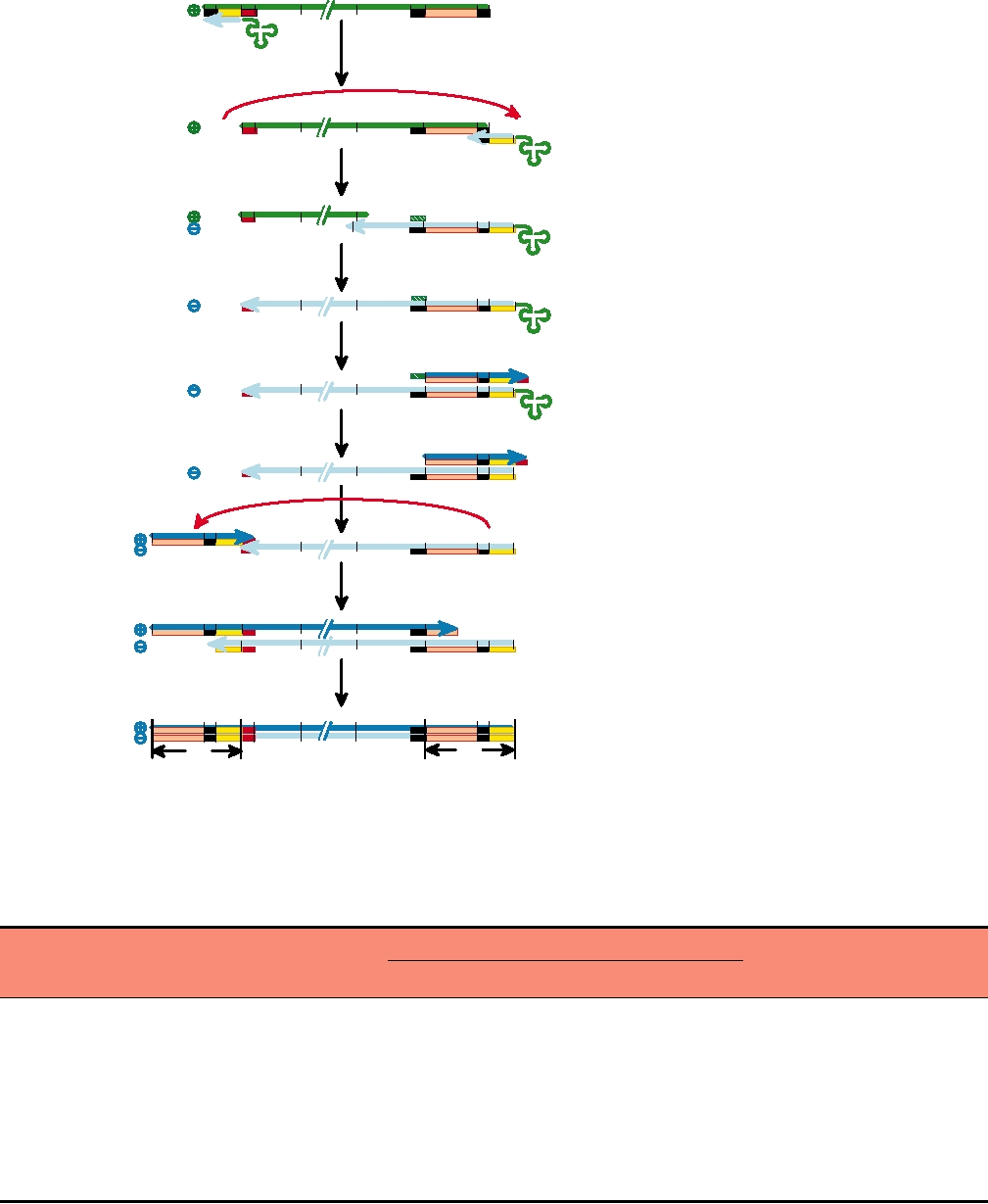
Synthesis of First DNA Strand
The process of reverse transcription of the viral genome
is illustrated in Fig. 6.3. At the ends of the viral RNA are
R
U5 PB
leader
PP
U3 R
domains that are essential for production of the dsDNA
copy of the RNA genome. These are illustrated schemati-
cally in Figs. 6.2 and 6.3, and the sizes of these elements are
gag
Genome
pol
env
CAP
An
shown in Table 6.2 for a number of different retroviruses.
A direct repeat element called R, 15230 nucleotides in
length depending on the virus, is present at the two ends of
the RNA genome. At the 5˘ end of the genome, a unique
Provirus
U3
U5
U3
U5
sequence element called U5 (70220 nt long) is present
LTR
LTR
immediately downstream of R, and at the 3˘ end of the
genome a unique element called U3 (2301200 nt) is present
Polymerase Envelope
Capsid
immediately upstream of R. On completion of DNA synthe-
proteins
proteins
sis, these elements give rise to direct repeats present at the
ends of the viral DNA, called the long terminal repeats or
Virion
LTRs, which have the sequence U3RU5.
Immediately 3˘ of U5 is a primer binding site (PBS), where
18 nucleotides are exactly complementary to the 3˘ end of a
specific cellular tRNA. Each genomic RNA molecule has
FIGURE 6.2 Diagram showing the overall organization of the genes
bound to it at the PBS one molecule of the appropriate tRNA,
in the retroviral RNA genome, the comparable organization of the DNA
which is used as a primer for DNA synthesis. The tRNA
provirus, and the location of the various virus-encoded proteins in the
mature virion. The RNA strand is shown in green; the DNA provirus is
used depends on the virus, and several different tRNAs are
shown in blue. The ORFs in the genome are color coded to match their
known to be used by different viruses (Table 6.2).
products in the virion. The nontranslated regions of the RNA genome and
Reverse transcription begins by extending the primer
the long terminal repeats (LTRs) are described in detail in the text. The pro
tRNA through U5 and R, and stops when the 5˘ end of the
gene, located between gag and pol, is not shown here. Adapted from Goff
RNA genome is reached (Fig. 6.3, steps 1 and 2). The cDNA
(1997), Figure 3.5 on p. 143.
59 R U5PBS gag pro pol
PPT
39
1) Primer tRNA anneals to PBS sequence
env
U3
R
An
in genome RNA
3' R' U5'
2) tRNA is extended to form DNA copy
of the 5' end of the genomic RNA
Strong Stop DNA
3) RNase removes hybridized RNA
(R and U5)
4) First Jump. DNA hybridizes to
An
remaining RNA R sequence at 3' end
R' U5'
PPT
U3'
R' U5'
5) DNA minus-strand extended and
completed; most RNA removed.
PPT
PPT
6) Plus-strand DNA primes at polypurine
track (PPT) downstream of env gene 5'
end of plus-strand DNA is synthesized
7) RNase H degrades tRNA and PPT
U39
R9 U59
PBS
8) Second Jump Plus-strand DNA binds
U39 R9 U59
U39
R9 U59
to the primer binding sequence (PBS)
near the 3 end of minus-strand DNA
9) Both strands extended and completed
to give double-stranded DNA with
duplicated LTRs in the same orientation
at both ends
PBS
PPT
59
39
gag pro pol
env
U3'
R U5
R U59
U3
LTR
LTR
39
59
FIGURE 6.3 Mechanism of retroviral DNA synthesis (reverse transcription). Green lines are RNA, light blue lines are
minus-strand DNA, and the dark blue line in the last three steps is plus-strand DNA. Features within the LTRs (U3, R,
U5), as well as the PBS and PPT (polypurine tract), are indicated by colored bars beneath the lines designating the nucleic
acids, to clarify what is present at each step. Adapted from Fields et al. (1996) p. 1792; Goff (1997) Figure 3.6 on p. 145;
Coffin et al. (1997) Figure 2 on p. 123.
TABLE 6.2 Terminal Regions of Retrovirus Genomes
Approximate sizes in bases of terminal elements
Genus
Prototype virus
U3
R
U5
Primer tRNA used
230a
Alpharetrovirus
RSV, ALV
20
80
Trp
1200b
Betaretrovirus
MMTV
15
120
Lys-3
Gammaretrovirus
MLV
450
70
80
Pro/Gln
Deltaretrovirus
HTLV-1
350
230
220
Pro
Epsilonretrovirus
WDSV
440
80
70
His
Lentivirus
HIV-1
450
100
80
Lys-1,2,3
Spumavirus
HRSV
910
190
160
Lys-1,2
a
Includes v-src gene.
b
U3 contains sag gene.
Source: Adapted from Table 2, page 38, in Coffin et al. (1997).
product, called the first strong stop DNA, is then transferred,
primers. Thus, the enzyme differs from the RNA repli-
while still attached to the primer, from the 5˘ end of the
cases of RNA viruses, which use other mechanisms for the
RNA to the 3˘ end ("first jump," Fig. 6.3, steps 3 and 4). It is
initiation of RNA replication
unknown whether transfer is usually to the 3˘ end of the same
molecule, to the 3˘ end of the second copy of the genome,
Why Is the Genome Diploid?
or randomly to either copy of the RNA in the virus. This
transfer uses the repeat element R at the 5˘ and 3˘ ends--the
Why the retroviral genome is diploid is not clear. In other
DNA copy detaches from R at the 5˘ end and anneals to R at
instances where reverse transcription occurs, such as in the
the 3˘ end, so that only one copy of R is present in the DNA
hepadnaviruses and the retrotransposons, the RNA to be cop-
transcript. RNase H is presumably important for this. During
ied is not diploid. Furthermore, in vitro studies have shown
reverse transcription, the RNA strand is destroyed by RNase
that retroviral RT can use a single copy of the RNA genome
H about 18 nt behind the transcription point. The degrada-
to produce a full-length dsDNA. Thus, two copies of the
tion of the RNA strand during transcription of the DNA copy
genome are not essential. However, it is possible that dur-
may encourage the jump to the other end. Other components
ing infection the process is more efficient if the RT can go
of the particle may also be involved in the jump.
back and forth between the two copies, and this resulted in
After the jump, reverse transcription resumes until the 5˘
selection for a diploid genome in retroviruses. The process of
end of the RNA template is reached. Note that the RNA strand
reverse transcription to produce the viral dsDNA is complex
now ends at PBS because RNase H has degraded the RNA
with the multiple jumps required, and the diploid genome
strand of the DNARNA hybrid (but not the RNA in PBS
could be organized in such a way as to make these jumps
because this is an RNARNA duplex). This process results
more efficient. Other possible advantages of a diploid genome
in the formation of what is called first-strand DNA or minus-
that could have resulted in selective pressure for diploidy are
strand DNA, because it is the antimessage sense (step 5).
the possibility of overcoming at least some damage in the
RNA by switching templates, and the fact that switching tem-
plates results in recombination. Recombination does occur
Synthesis of the Second DNA Strand
frequently in retroviruses, and it is clear from many studies
The first-strand DNA produced in this way is the tem-
that recombination has been important in their evolution.
plate for second-strand (plus-strand) DNA synthesis. The
primer for second-strand synthesis is an RNA oligonucle-
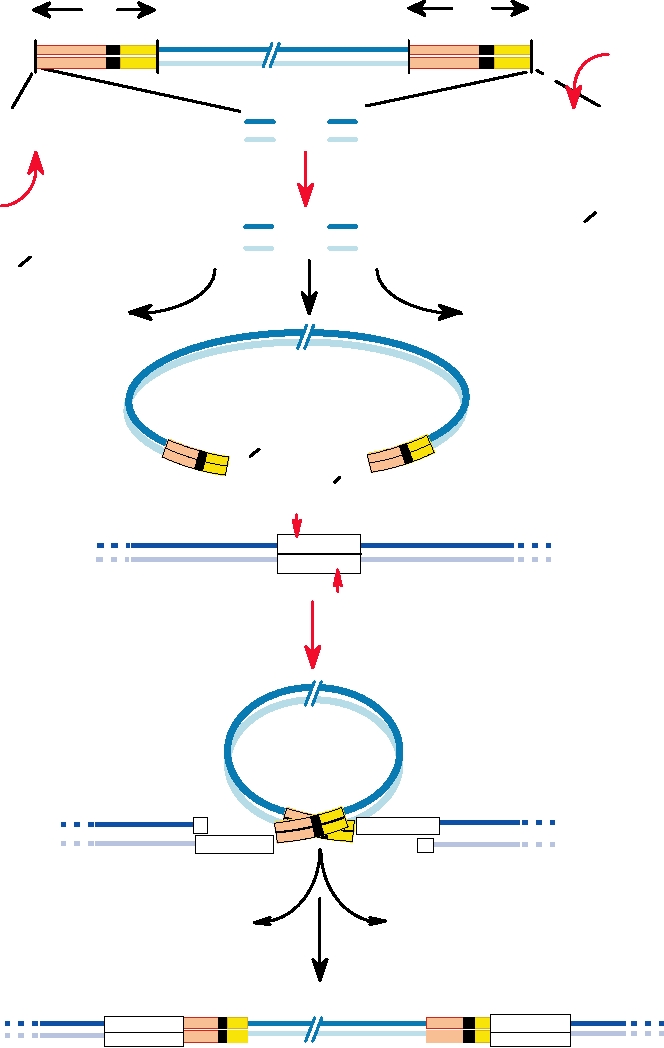
Integration
otide, called the polypurine tract or PPT, positioned imme-
diately 5˘ of U3. PPT survives RNase H degradation and its
After the appearance of the full-length dsDNA genome
3˘ terminus is exact; that is, the cleavages to produce it are
in the nucleus, the viral integrase catalyzes its insertion into
precise (Fig. 6.3, step 6). In addition to this precise primer
a host cell chromosome by means of a single recombina-
for plus-strand synthesis, which defines the boundary of U3
tional event. This process is illustrated in Fig. 6.4. Insertion
and thus of the LTR, additional priming sites are used in
is essentially random within the host genome. The first event
in integration is the removal of two nucleotides from the 3˘
some retroviruses, such as HIV.
The PPT plus-strand primer is extended through U3, R,
end of both strands of the viral DNA. The next two nucle-
otides are always AC, from 3˘ to 5˘. The 3˘ OH of the now
U5, and into the region of the primer tRNA, which is still
3˘-terminal A residue is used to attack an internucleotide
attached to the first-strand DNA. The 18 nucleotides of the
tRNA primer that are complementary to the PBS are copied,
phosphate in the host DNA. Attack is coordinated so that
but further copying is thought to be blocked by a modified
both ends of the viral DNA are inserted at once. The spac-
nucleotide in the tRNA that cannot be copied. The tRNA
ing between the two insertion points depends on the viral
primer is then removed by RNase H (step 7) and a second
integrase and is 4, 5, or 6 nucleotides in different viruses.
jump occurs. In the second jump, the nascent second-strand
The ends of the inserted structure are then cleaned up, prob-
DNA is transferred to the other end of the template, using
ably by host enzymes. Insertion results in the loss of the two
the PBS sequence, which is now present at the 3˘ ends of
terminal nucleotides of the viral DNA. In addition, a duplica-
both strands (second jump, step 8). Synthesis of both strands
tion of the 4, 5, or 6 nucleotides of the host that lie between
of DNA resumes and it becomes full length and double
the two insertion points is produced, and these duplicated
stranded. The resulting dsDNA is cleaned up, probably by
nucleotides flank the two ends of the viral genome.
cellular enzymes. In the full-length, linear ds copy of the
The integrated form of the virus, the provirus, is stable.
genome, the LTR sequence U3RU5 has been formed at
No mechanism for precise excision of the provirus is known,
both ends of the DNA genome (Fig. 6.3, step 9).
and integration is essentially irreversible. Most retroviruses
It is of interest that RT, like DNA polymerases, requires
do not kill the host cell, and the provirus behaves as a simple
a primer to synthesize DNA, but unlike most DNA polymer-
Mendelian gene that is transmitted to all daughter cells. It
ases it can copy either DNA or RNA, given the appropriate
is obvious that insertion of such a provirus into the germ
LTR
LTR
5'
3'
U3
R U5
U3
R U5
Nucleophilic
5'
attack by water
3'
TGTGGAAAATCTCTAGCAGT
ACTGGAAGGGCTAATTCAC
ACACCTTTTAGAGATCGTCA
TGACCTTCCCGATTAAGTG
Integrase
End processing
Nucleophilic
3' H
O
attack by water
TGTGGAAAATCTCTAGCA
ACTGGAAGGGCTAATTCAC
ACACCTTTTAGAGATCGTCA
ACCTTCCCGATTAAGTG
OH
3'
GT
TG
Provirus
3'
OH
ACTG
CA
AC
GTCA
OH
3'
X1234Y
Host DNA
Y 4 3 2 1 X
Integrase
Joining reaction
AC
X
CA1234Y
TG
GTCA
1 2 3 4 X
AC
Y
Cellular enzymes
Repair
AC
CA
Integrated provirus
U3 R U5
U3 R U5
X1234TG
CA1234Y
Y 4 3 2 1
4 3 2 1 X
GT
AC
FIGURE 6.4
Steps in the integration of retroviral DNA into the host genome. First, the termini of the blunt-ended
viral DNA are attacked by the integrase, and two bases (blue) adjacent to a highly conserved CA dinucleotide (red) are
lost by nucleophilic attack by a water molecule, leaving recessed 3˘ OH ends. Next, the host DNA is cleaved at the target
sequence and the 3˘ OH ends of the viral DNA are joined by the integrase to the 5˘ phosphates on the host DNA. Overhang
removal and gap and nick repair by cellular enzymes complete the integration reaction. Adapted from Hindmarsh and
Leis (1999); Goff (1997) Figure 3.10 on p. 145, and Coffin et al. (1997) Figure 8 on p. 185.
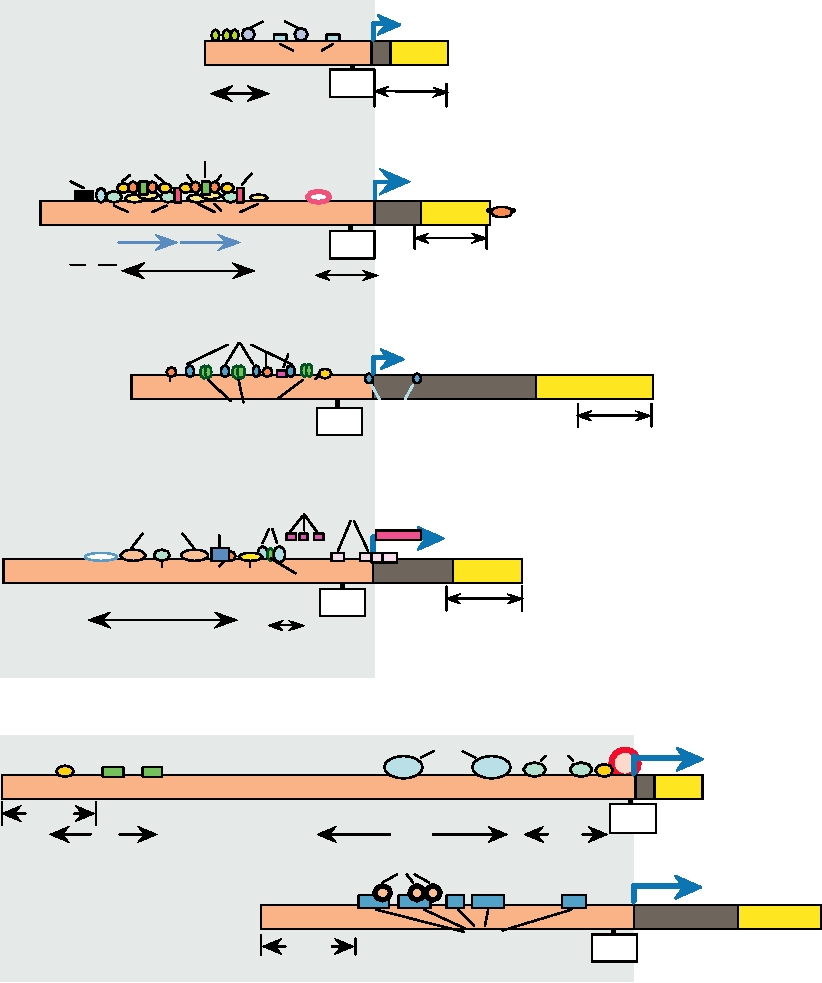
line would lead to its transfer to progeny organisms, which
polymerase II. The transcription signals, almost all found in
appears to have occurred many times in the past.
U3, include TATA boxes as well as an array of binding sites
for cellular transcription factors that are optimal for the cell in
which the particular retrovirus primarily replicates. Fig. 6.5
Transcription of RNA in Simple Retroviruses
gives examples of the binding sites for transcription factors
The LTR of the integrated provirus has all of the signals
in several different retroviruses. This figure illustrates that
required for transcription of the provirus by cellular RNA
the constellation of binding sites is complex and is different
U3
R
U5
EFII
EFI
RSV
EFIII
enhancer
TATA
100bp
CBF
bHLH
ELP NF-1 ETs ETs
UCRBP
C/EBP
Factor A
MLV
MCREF
GR
TATA
100bp
PBS
enhancer
promoter
() ()
()
(+)
(+)
Myb ETs Sp-1
PTLV-1
NF-1
ETs
Myb
21 bp TREs
TATA
100bp
Sp-1 LBP-1 TAR
NF-κB
NFAT-1 USF-1
COUPTF
HIV-1
AP2
ETs TCF-1
GR
enhancer TATA
100bp
NRE
(+/)
(+)
U3
R
U5
Oct 1
NF-1
MAF
NBP
GR
MMTV
NF-1
200bp
TATA
ME
NRE
HRE
AP1
HSRV
BREs
200bp
TATA
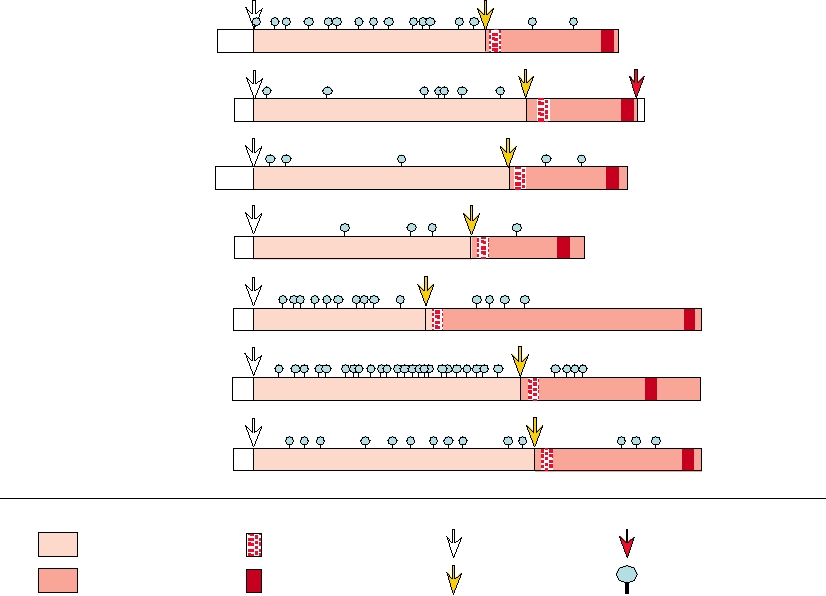
FIGURE 6.5 Transcription signals in retroviral LTRs. Where the same factor binds to a number of LTRs, the same
symbol has been used. The TRE elements in PTLV-1 are the Tax-responsive elements. The blue arrow marks the
beginning of transcription. Note that the scale of the lower panel is twofold different from that in the upper panel.
Composite of Figures 4, 5, 7, 8, 9, 11 in Chapter 6 of Coffin et al. (1997).
in different viruses that replicate in different tissues. Only
rovirus MasonPfizer monkey virus, this element is called
the upstream (5˘) LTR is used to initiate transcription--or at
the constitutive transport element (CTE). It is 154 nucle-
otides long and is located in the 3˘ nontranslated region of
least efficient transcription.
Transcription initiates precisely at the 5˘ end of R, as
the RNA. In the avian retroviruses, an apparently unrelated
element of about the same size, also present in the 3˘ non-
indicated by the arrow in Fig. 6.5, and proceeds through
the proviral genome and into the 3˘ flanking sequences. The
translated region, provides the same function for unspliced
transcript is capped by host cell capping enzymes. There is
avian retroviral RNA. Interestingly, the avian sequence does
a poly(A) addition signal (AAUAAA) in the RNA, which is
not work in mammalian cells. The monkey virus sequence
usually present about 2030 nucleotides upstream of the end
works in both mammalian and avian cells, but works better
of the viral genomic RNA sequence (i.e., upstream of the 3˘
in mammalian cells. These findings are consistent with the
end of R). Host cell polyadenylation machinery recognizes
hypothesis that these transport elements interact with cellu-
the AAUAAA signal, cleaves the RNA transcript precisely
lar proteins. The inability of unspliced RNA to be exported
at the RU5 boundary, and polyadenylates the RNA. Thus
from the nucleus is one of the reasons that the avian retrovi-
the processed RNA transcript is identical to the genomic
ruses will not replicate in mammalian cells.
RNA and a round-trip from RNA to DNA to RNA has been
made.
Translation of Viral Genomic RNA
In many viruses, the polyadenylation signal is present in
Retroviral genomic RNA is translated into two or three
U3. Thus, there is only one copy of this signal in the RNA
transcript, near the 3˘ end. However, in some viruses the sig-
polyproteins that are eventually processed by the viral
nal is present in R and is therefore present in both the 5˘ and
protease. The order of genes along the genomic RNA is
3˘ regions of the transcript. Interestingly, only the 3˘ copy of
gagpropol, encoding the proteins Gag, Pro, and Pol. Stop
codons are present between Gag and Pro, or between Pro and
the signal is active for cleavage and polyadenylation of the
Pol, or in both places. Termination of the polypeptide chain
transcript.
occurs at these stop codons most of the time during transla-
Some fraction of the genomic RNA is exported to the
tion. These stop codons are suppressed some of the time,
cytoplasm without further processing. There it serves as
however, either by readthrough or by frameshifting (Chapter
mRNA for the synthesis of Gag, GagPro, and/or GagPro
1), so that the amount of Pol produced is usually about 5%
Pol, as described later. Alternatively, it can be packaged
that of Gag. In viruses with one stop codon, the frequency
into progeny viruses. Genomic RNA that serves as mRNA
of suppression is about 5%, but in viruses with two stop
and genomic RNA that serves as the source of RNA for
codons, the frequency of suppression of each stop codon is
packaging are maintained in separate pools.
higher so that significant amounts of Pol are produced even
Some fraction of the genomic RNA is spliced before
though suppression of two stop codons is required. This
export to the cytoplasm. Only one spliced RNA is made in
means that the frequency of suppression is variable and can
the simple retroviruses, which serves as mRNA for Env. In
be controlled by changes in the sequence of the viral RNA.
most retroviruses, the entire GagProPol region is spliced
Because reinitiation does not occur once the chain is termi-
out and the initiation codon for Env is encoded in env. In
nated, the polyproteins produced are Gag and/or GagPro
the avian retroviruses, however, the upstream splice site is
and/or GagProPol, depending on the positions of the stop
located within the Gag coding sequence so that Env begins
codons (Fig. 6.6).
with the first six codons of Gag.
Pro is produced in three different ways in different retro-
The need for both spliced and unspliced versions of the
viruses, as illustrated in Fig. 6.6. In the avian viruses, such
viral RNA means that mechanisms must exist to ensure that
as ALV, there is no stop codon between Gag and Pro so
both are produced and that the ratio of spliced to unspliced
that a GagPro polyprotein is produced. Gag and Pro are
RNA is optimal for virus replication. In the simple retrovi-
thus produced in equal amounts. Frameshifting results in
ruses, the splice sites are suboptimal, so that not all RNA is
the production of a longer polyprotein, GagProPol. Most
spliced. Experiments have shown that the result is an opti-
of the mammalian viruses also only have a single stop
mal ratio of spliced to unspliced RNA. Mutations that make
codon in the ORF, but it is positioned between Gag and
splicing more efficient are deleterious for virus growth and
Pro. Readthrough of a UAG stop codon (murine leukemia
revertants quickly arise that restore the proper ratio. The
viruses) or frameshifting (other mammalian viruses with
second problem faced by these viruses is the need to export
a single stop codon) results in the longer polyprotein. The
unspliced RNA to the cytoplasm. Eukaryotic cells have
two polyproteins produced are Gag and GagProPol, and
control mechanisms to ensure that RNA containing splice
Pro and Pol are produced in the same low amounts. Finally,
sites is not exported from the nucleus. It has been found
several mammalian retroviruses, for example, MMTV and
that sequence elements in the unspliced RNA are required
PTLV-1, have two stop codons in the ORF, both of which
for its export, and it is assumed that these elements interact
can be suppressed by frameshifting. Thus, three polyproteins
with cellular proteins that promote export. In the simian ret-
GAG
PRO
POL
Alpharetrovirus (ALV)
RT
MA
CA
NC Pro
IN
Pr76 gag-pro
p95 (β)
Pr180 gag-pro-pol
(5%)
Gammaretrovirus (MLV)
Pr65 gag
Pr180 gag-pro-pol
(5%)
Betaretrovirus (MMTV)
Pr77 gag
Pr110 gag-pro
DU
Pr160 gag-pro-pol
(25%)
(10%)
Deltaretrovirus (PTLV-1)
Pr55 gag
Pr70 gag-pro
Pr165 gag-pro-pol
Epsilonretrovirus (WDSV)
Pr gag
Pr gag-pro-pol
Lentivirus (HIV)
Pr55 gag
p66
Pr165 gag-pro-pol
(10%)
Spumavirus (HSRV)
Pr78 gag
Pr125 pro-pol
Pro Protease
RT Reverse transcriptase
Frameshift
DU UTPase
IN Integrase
Readthrough
RNase H (where known)
FIGURE 6.6 Organization of the gag, pro, and pol genes of representative retroviruses belonging to each genus. In
some cases two frameshifts are required to generate a complete GagProPol precursor. The gag proteins are illustrated
in more detail in Fig. 6.7. This figure is a composite of Goff (1997) Figure 3.16 on p. 157; and Coffin et al. (1997) pp. 45,
269, 795, and 799. Virus name abbreviations can be found in Table 6.1.
are produced, Gag, GagPro, and GagProPol. In this case
site. Because the monomer is not active, there is a delay in
Pro is produced at intermediate levels.
processing. The high concentration of viral polyproteins that
Processing of these various polyproteins occurs during
occurs in viral particles or in previral particles is required to
assembly of progeny virions. The viral Pro is an aspartate
achieve efficient dimerization of the protease and its activa-
protease whose active site contains two aspartic acid residues
tion. Experiments have shown that premature activation of
(Chapter 1). The protease domain is functional in polypep-
the protease, which can be achieved by using genetic tricks,
tides containing the Pro sequence as well as after its release
is deleterious for virus assembly. Thus, it is important that
by proteolysis as a small protein of about 100 residues. The
processing be delayed until assembly begins or is completed.
enzyme is active only as a homodimer, with each chain in
During processing of Gag, several different proteins are
the dimer supplying one of the aspartic acids in the active
produced, some of which are quite small, whereas others are
a homodimer of RT. In other viruses, cleavage between RT
larger. The proteins produced from Gag are illustrated sche-
and IN is incomplete, and the active enzyme is a heterodimer
matically in Fig. 6.7 for a number of retroviruses. Gag is
of RT and RT-IN. A third pattern occurs in HIV, in which
cleaved to produce at least three protein products in all ret-
partial cleavage occurs between the polymerase domain and
roviruses except the spumaviruses, called MA (membrane-
the RNase H domain. In this case, the products of Pol are a
associated or matrix protein), CA (capsid protein), and NC
truncated RT lacking the RNase H domain (called p51), the
(nucleocapsid protein). These three peptides are always
full-length RT (called p66), and IN. The active reverse tran-
present in that order from the N terminus to the C terminus
scriptase is a heterodimer between p51 and p66.
in the Gag polyprotein. Gag (and thus MA) is myristoylated
in most retroviruses and associates with membranes.
Myristoylation may serve to recruit Gag to membranes for
Production and Processing of Env
assembly or budding. NC is a small basic protein that binds
RNA and Zn2+. It has a number of functions, including bind-
Translation of the mRNA for Env produces the envelope
ing to the packaging signals in the viral RNA that lead to
glycoprotein precursor. Processing of Env and the location
its incorporation into virions, the facilitation of binding of
of key features are illustrated in Fig. 6.8 for a number of ret-
the tRNA primer to the genomic RNA, the formation of
roviruses. The precursor has an N-terminal signal sequence
the genomic RNA dimer, and strand transfer during reverse
(labeled L in the figure), which leads to its insertion into
transcription. CA is a larger protein that is believed to form
the endoplasmic reticulum, a membrane anchor sequence
a shell around the viral RNA and its associated internal pro-
near the C terminus, and a C-terminal cytoplasmic domain
teins (Fig. 2.21). In addition to these three proteins, in most
(i.e., the protein is a type I integral membrane protein). The
retroviruses other proteins, whose functions are not well
signal sequence is removed during translocation into the
understood, are also produced from Gag (Fig. 6.7).
endoplasmic reticulum, and the protein is glycosylated and
Processing of Pol varies in different viruses. Three pat-
transported to the plasma membrane by conventional cel-
terns of cleavage can be distinguished. In some viruses,
lular pathways. As with many other viral glycoproteins, the
RT, consisting of the polymerase domain and the RNase H
precursor is cleaved by furin during transport to produce an
domain, is released from the upstream Pro and the down-
N-terminal extracellular component called SU (for surface)
stream IN. The active reverse transcriptase is a monomer or
and a C-terminal membrane-spanning component TM (for
CA (capsid)
NC (nucleocapsid)
MA (matrix)
99Minimum99
GAG
ALV
p12
p19
p27
p10
MLV
p15
p12
p30
p10
MMTV
p10
p27
p21
p3 p8?
p14
PTLV-1
p24
p15
p19
WDSV
p10
p25
p14
p20
(p10)
HIV-1
p7
p6
p24
p17
HSRV
(CA)
(MA)
(NC)
Myristate
Acetate
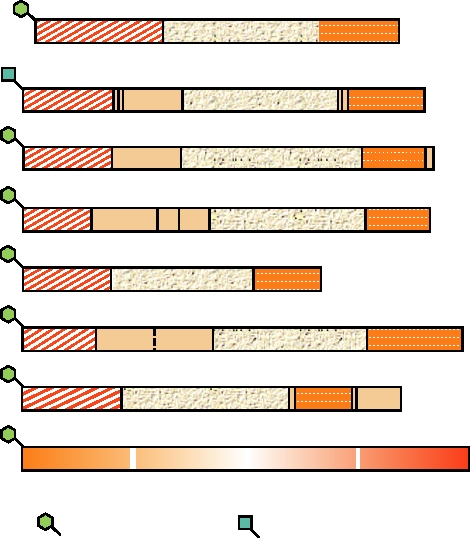
FIGURE 6.7 Organization of the Gag proteins in representatives of each retroviral genus. The viruses are shown in the
same order as those in Fig. 6.6. Vertical solid lines mark sites of cleavage by the viral protease. Sequences representing
the mature matrix, capsid, and nucleocapsid proteins are indicated with different shadings, and the approximate molecular
weights of the processed proteins are shown. Note that the Gag polyprotein of HSRV is not processed. From Coffin et
al. (1997) pp. 44 and 798.
ALV
L
gp85
gp37
MLV
p15E/12E
L
gp70
MMTV
L
gp52
gp36
PTLV-1
gp46
gp21
L
WDSV
L
SU
gp90
HIV
L
gp120
gp41
HSRV
gp80
gp48
L
Fusion domain
SU protein
Signalase
Pro (viral protease)
Transmembrane
TM protein
Furin
N-linked carbohydrate
anchor
FIGURE 6.8 Organization of the envelope proteins of representatives of each genus of retroviruses, shown in the
same order as in Figs. 6.6 and 6.7. The domains corresponding to the mature SU and TM proteins are shown in different
shades of color. Cleavages by signalase to remove the leader peptide (L) are indicated with white arrows, and those due
to furin by yellow arrows. The red arrow marks the site of cleavage in MLV by the viral protease. The fusion domains,
the transmembrane domains, and the sites of predicted N-linked carbohydrate addition are shown. Adapted from Coffin
et al. (1997) Figure 10 on p. 56. Virus name abbreviations can be found in Table 6.1.
transmembrane). SU and TM remain associated. In some
The accessory genes of the complex retroviruses are
cases, the association is stabilized by disulfide bonds, but
located upstream or downstream of env and are translated
in other cases there is no covalent linkage. SU is always
from spliced mRNAs, with some of the genes requiring
glycosylated, whereas TM may or may not be glycosylated.
multiple splicing for expression. The genome organizations
Cleavage to produce SU and TM is required for the activation
of the different genera are diagrammed in Fig. 6.9 to show
of the fusion activity, which is located near the N terminus
the location of these accessory genes. The different acces-
of TM. Thus, the production of the envelope glycopro-
sory genes have been inserted into the retroviral genome at
teins of the retroviruses parallels that of the envelope gly-
different times during the evolution of these viruses (Fig.
coproteins of many enveloped RNA viruses.
6.1), and insertion of such genes appears to be a dynamic
process that is ongoing. As one example, the vpx gene of
HIV-2 and the vpr gene of HIV-1 appear to have been
Accessory Genes of Complex Retroviruses
inserted into their respective viruses after the separation of
In addition to the core genes gag, pro, pol, and env that
HIV-1 and HIV-2.
are present in all retroviruses, members of the four genera of
The presence of the accessory genes allows more vigor-
complex retroviruses possess additional genes that allow them
ous replication of the retrovirus that possesses them, which
to regulate the development of the infection cycle. Accessory
can be fatal to the host cell. As described before, the simple
genes are also present in the betaretroviruses, but these are
retroviruses do not kill the host cell, but instead establish a
not regulatory in nature. A listing of the accessory genes in
persistent infection in which the cell survives and produces
different retroviruses is given in Table 6.3, and a complete
low levels of virus indefinitely. However, at least some of
listing of all of the proteins of HIV is shown in Table 6.4.
the complex retroviruses, such as HIV-1, can replicate to
TABLE 6.3 Accessory Genes in Retroviruses
(2) activation of transcription of the provirus to greatly
increase the rate of production of viral RNA; (3) export of
Gene
Functions
unspliced viral RNA to the cytoplasm; (4) arrest of the cell
cycle in infected T cells; (5) promotion of virus assem-
Betaretrovirus (MMTV)
bly and release; and (6) protection from cellular defense
sag
Superantigen
mechanisms.
dut
dUTPase
Deltaretrovirus (HTLV/BLV)
Transport of Viral DNA into the Nucleus
tax
Transcription activator (like tat)
rex
Splicing/RNA transport regulator (like rev)
HIV-1, and probably other lentiviruses as well, can infect
Lentivirus (HIV-1)
quiescent cells because the viral DNA and associated pro-
tat
Transcription activator (like tax)
teins can cross the nuclear membrane. The product of the vpr
rev
Splicing/RNA transport regulator (like rex)
gene appears to be required for this. MA and IN have also
been implicated in the process.
vif
vpr/vpx
See Table 6.4
nef
Transactivation of the Transcription of Viral RNA
vpu
All complex retroviruses encode a protein that transacti-
dut
dUTPase (in nonprimate lentiviruses)
vates transcription of viral RNA from the provirus. In PTLV/
Facilitates replication in certain cell types.
BLV, the gene for this protein is called tax. The Tax protein
Spumavirus (HSRV)
activates transcription by means of sequence elements in the
tas (bel 1)
Transcription activator (DNA binding protein)
U3 region of the viral DNA called Tax-responsive elements
bel 2
?
(TREs), whose locations are shown in Fig. 6.5. Tax interacts
bet
? (may be involved in latency)
with cellular transcription factors that bind to TREs, and this
Epsilonretrovirus (WDSV)
interaction results in increased activity of the transcription
factors. One such transcription factor is the cAMP response
Orf A
Viral homologue of cyclin D
element/activating transcription factor (CREB/ATF). Tax
Orf B
?
also increases the transcription of certain cellular genes, in
Orf C
?
some cases through its interactions with CREB/ATF, and in
other cases by stimulating the transcription of genes regu-
Source: Adapted from Coffin et al. (1997) Table 1 on p. 36, and
lated by NFκB. Many of these cellular genes are important
information from Fauquet et al. (2005).
in the regulation of T cells, and their stimulation may relate
high titer in some cell types with the result that the cells
to the pathology of disease caused by the virus.
die. It is interesting that many endogenous retroviruses
In the lentiviruses, the gene for the transcriptional acti-
are present in the germ line of different vertebrates, as
vation is called tat. The Tat protein works in one of two
described later. However, none of these are complex ret-
different ways. Tat of visna virus appears to interact with
roviruses. It is possible that the regulated lifestyle of the
cellular transcription factors in a manner similar to Tax,
complex viruses would make complex endogenous viruses
using a sequence element in U3, although the cellular fac-
difficult to silence. The inability to silence such viruses
tors are different. Tat of HIV and its close relatives, how-
could lead to selection against organisms that contain them
ever, stimulates transcription by binding to a sequence
in the germ line.
element called TAR at the 5˘ end of the viral RNA. TAR is
Many of these proteins encoded by the accessory genes
composed of the first 60 nucleotides of HIV-1 RNA, which
are multifunctional and their functions are only partially
form a stem-loop structure that is essential for the function
understood. The intense interest in PTLV and HIV has
of TAR. TAR of both HIV-1 and HIV-2 are shown in Fig.
resulted in extensive study of their accessory genes, tax
6.10. How the binding of Tat to TAR activates transcrip-
and rex in the case of PTLV/BLV and tat, rev, vif, vpr/
tion is not yet clear. One model is that Tat interacts not
vpx, nef, and vpu in the case of lentiviruses. The acces-
only with the nascent viral RNA but also with cellular tran-
sory genes of the spumaviruses, bell, bel2, and bet, as well
scription factors, and in so doing stabilizes the transcrip-
as those of the epsilonretroviruses, have been less well
tion complex or changes its composition. In this model, the
studied. The functions of these genes can be conveniently
altered transcription complex is more processive, allow-
grouped into six categories: (1) transport across the nuclear
ing the production of complete RNA genomes rather than
membrane of subviral particles that synthesize viral DNA
truncated transcripts. It may also initiate transcription more
after infection, allowing the virus to infect quiescent cells;
frequently.
TABLE 6.4 The HIV Proteins
Protein
mRNA
Size (kD)
Post-translational modifications
Functions
gag
Genomic RNA
p25 (CA)
None
Capsid structural protein
p17 (MA)
Myristoylated at Gly-2
Matrix protein
p7
?
RNA-binding protein
p2
?
RNA-binding protein
pro
Genome RNA
p10 (PR)
Viral protease, processes gag proteins
frameshifted
pol
Genome RNA
p66/p51 RT
Heterodimer, p51 lacks RNase
Reverse transcriptase
frameshifted
H domain present in p66
vif
vif mRNA
p23
Viral infectivity factor, essential for spread in macrophages
vpr/vpx mRNAa
vpr/vpx
p15
Associates with p7
Augments replication
tat
tat mRNA
p14
Required for replication transactivates RNA synthesis,
binds to TAR RNA
rev
rev mRNA
p19
Regulates splicing/RNA transport; binds RRE element and
facilitates env translation
vpu
vpu/env mRNA
p16
Phosphorylated on Ser
Helps in virion assembly and release, dissociates gp
160/CD4 complex
env
vpu/env mRNA
gp 120 (SU)
24 sites for N-linked glycosylation
Surface glycoprotein, mediates cellular attachment
gp 41 (TM)
7 sites of N-linked glycosylation
Transmembrane glycoprotein
nef
nef mRNA
p27
Myristoylated at Gly-2,
Homodimer, causes pleiotropic effects, including
phosphorylated at Tyr-15
downregulation of CD4
a
vpr is found in HIV-1; vpx is found in HIV-2.
Source: Adapted from Levy (1994) Table 1.4, p. 8.
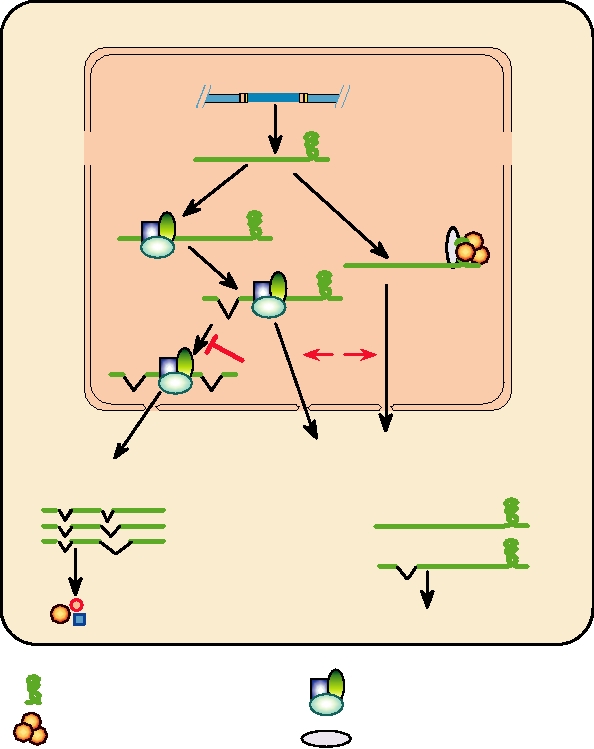
Export of Unspliced Viral RNA to the Cytoplasm
lular proteins involved with the nuclear export pathway.
Complex retroviruses encode proteins that promote
The end result is that Rev promotes the export of RNAs
the export of unspliced or partially spliced RNA from the
containing RRE (i.e., unspliced genomic RNA and singly
nucleus. The proteins are Rex in the case of PTLV/BLV and
spliced mRNAs) from the nucleus to the cytoplasm (Fig.
Rev in the case of lentiviruses. Rex and Rev are translated
6.12). Rev appears to accompany the RNA to the cyto-
from multiply spliced mRNAs, as are the transcriptional acti-
plasm and then to cycle back to the nucleus. Thus, after the
vators and Nef in the case of the lentiviruses. The multiple
appearance of Rev, the infection cycle switches to a late
splicing events in HIV-1 are illustrated in Fig. 6.11. Early
phase with the appearance in the cytoplasm of the mRNAs
in infection, before Rex (PTLV/BLV) or Rev (HIV-1, -2)
for GagProPol, Env, Vpu, Vif, and Vpr. Genomic RNA
are present, only completely spliced mRNAs are exported
exported to the cytoplasm can also be packaged into
from the nucleus to the cytoplasm. Thus, the proteins made
progeny virions.
early are the transcriptional activators, Nef, and the proteins
In addition to its function in the export of viral RNAs
that control the export of unspliced or partially spliced viral
to the cytoplasm, Rev also has other functions. These func-
RNAs from the nucleus. The transcriptional activators accel-
tions appear to include regulation of the splicing of HIV
erate the production of viral RNAs, and Rex and Rev allow
RNAs and increasing the efficiency of translation of HIV
the export of mRNAs for the other viral proteins, which
RNAs (Fig. 6.12).
includes the genomic RNA. These processes are illustrated
The PTLV Rex appears to function similarly to Rev. It
schematically in Fig. 6.12.
binds to an RNA element, called the RexRE, that is similar
Studies with HIV-1 have shown that Rev binds to a
in size to the RRE, and binding of Rex promotes export of
sequence element in the viral RNA called the Rev response
viral RNA from the nucleus. However, the RexRE is found
element or RRE. HIV-1 RRE is 234 nucleotides in size and
in the U3R region of the genome and is therefore present
has multiple stem-loop structures that are important for
in all of the viral mRNAs. It is curious that Rex binds HIV-1
function. It is found in the env gene region and is therefore
RNA, although not in the same place as Rev, and will sub-
spliced out of the multiply spliced mRNAs (Fig. 6.11). In
stitute for Rev in HIV-1. However, Rev does not bind PTLV
addition to binding to RRE, Rev also interacts with cel-
RNA and will not substitute for Rex.
Alpharetrovirus
ALV
Gammaretrovirus
MLV
Betaretrovirus
MMTV
orf
Deltaretrovirus
PTLV
tax
rex
Epsilonretrovirus
WDSV
a
c
b
Lentivirus
HIV
vif
vpr
nef
tat
rev
vpu
Spumavirus
HSRV
bel1 bel3
bel2
bet10kb
0
2
4
6
8
R
GAG
POL
U3 U5
LTR
ENV
PRO
FIGURE 6.9 Coding regions of representatives of each retrovirus genus. Sites of translation initiation are shown with
arrows. The locations of the four major genes are shown in different colors. Accessory genes are named. Redrawn from
Coffin et al. (1997) Figure 5 on p. 37, and from Fields et al. (1996) p. 1776.
Cell Cycle Arrest in T Cells
interact with Gag. They are found in the virion in amounts
similar to that of Gag. Vif is a membrane-associated protein
HIV-1 kills infected T cells during active replication by
that is also present in the virion, but in amounts similar to
arresting the cell cycle. Cell death results from necrosis,
Pol rather than Gag. It has an effect on the processing of
not apoptosis as in the case for many viruses. The result in
Gag and is required for the production of infectious virions
infected humans is the depletion of T cells and, ultimately,
as described later. Nef, found in the primate lentiviruses,
progression to AIDS. The products of the vif and vpr genes
is a multifunctional protein that has a role in the assem-
are required for this arrest. Elimination of both genes, but
bly and release of infectious virus. Myristoylated forms
not of either one separately, results in a virus that cannot
of Nef are associated with the virion, where they may be
cause cell cycle arrest.
required for full infectivity of the virion. Another function
of Nef is the removal of CD4, the receptor for HIV, from
the surface of the infected cell. Removal of CD4 prevents
Assembly Functions
the interaction of Env with CD4 and facilitates the assem-
Several accessory proteins of the lentiviruses promote
bly and release of viruses. It also prevents superinfection of
the assembly and release of infectious virus, either directly
the cell by released virus. Vpu, a small integral membrane
or indirectly. Vpr, found in most lentiviruses, including
protein found in HIV-1 but not in HIV-2, promotes virus
HIV-1, and Vpx, found in HIV-2, are structural proteins that
release in two ways. It promotes the degradation of CD4 by
U G
C
70
G
GG
G
G
G
A
G
40
U
G
G U
30
U
U
A
C
A
U
U
60
C
G
C
C
G
C
A
U
C
C
U
G
C
30
U
U
U A CU
G
C
U G
50
C
U
A
C
80
40
U
A
U
A A
A
G
G
C
G
G
U
C
20
A
U
C
20 GG CA
G
C
G
C
A
A U
G
C
G C
A
U
A U
100
G C 90
U
A
A
U
A
U
CCAG CAC U G
G
C
G
50
G
G
U
10 U
C GGGUCGUGGC C
A
G
C
10 G C 110
G
U
U A
G
C
C G
U
A
U A
C
C
U
A
G C
G
C
C G
G
C
U G 120
60
1 G
C
G C
130
59
ACUGCUUA. . .
1 G C
C
59
CACGCUUGCUUGCUU. . .
HIV-1 TAR
HIV-2 TAR
FIGURE 6.10 Structures of HIV-1 and HIV-2 TAR RNA elements. Nucleotides are numbered from the 5˘ end of the
RNA. For HIV-1, positions involved in binding to Tat protein (yellow oval) are circled, and the bases involved in tertiary
structure alterations following Tat binding are shown in red. Less is known about the HIV-2 Tat binding. Adapted from
Coffin et al. (1997) Figure 12 on p. 226.
LTR
LTR
Proviral DNA
tat
rev
vpu
nef
Frame 3
pro
vpr
Frame 2
ORFs
env
pol
Frame 1
gag
vif
rev
tat
RRE
TAR
Genomic RNA
(gag mRNA) CAP
An
(gag-pro-pol mRNA)
Other mRNAs
An
vif mRNA CAP
An
vpr mRNA CAP
An
tat mRNA CAP
An
rev mRNA CAP
An
vpu, env mRNA CAP
nef mRNA CAP
An
1
2
3
4
5
6
7
8
9
kb
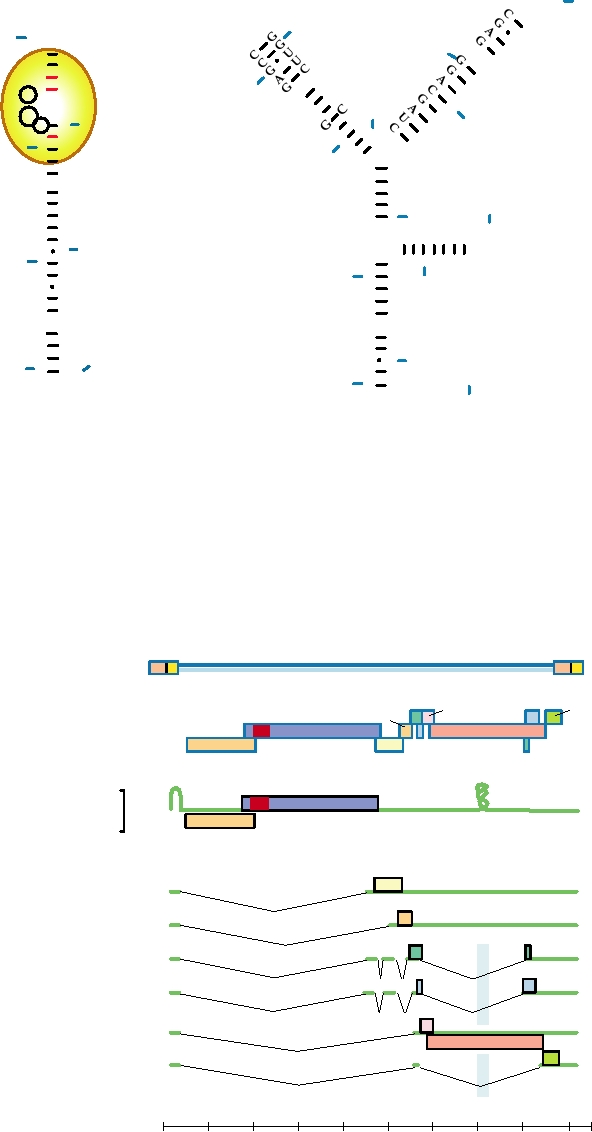
FIGURE 6.11 Genome organization and transcription map of HIV-1, the human immunodeficiency virus. The genome
is shown on the top line as the integrated provirus. The LTRs and all open reading frames (ORFs) are indicated. Below
this, the unspliced genome RNA is shown, with TAR and RRE (the Rev response element in env) indicated. The various
spliced mRNAs (and the ORFs translated from them) are diagrammed below the RNA genome. The pale blue shading
indicates the location of the RRE, which is spliced out of tat, rev, and nef messages. Redrawn from Coffin et al. (1997)
p. 803.
EUKARYOTIC HOST CELL
NUCLEUS
Integrated provirus
of complex
retrovirus
RRE
Early events
Late events
(minus REV)
(plus REV)
An
Spliceosome
+ Rev oligomer
+RAB
An
An
An
Further
splicing
B
A
An
Export of
Export of
non- and singly spliced mRNAs
multiply spliced mRNAs
An
An
An
An
An
Translation
Translation
C
Rev, Nef, Tat
Gag, Pro, Pol, Env, and other proteins
REV response element (RRE)
Components of spliceosome
Rev oligomer
RAB (similar to nucleoporins)
FIGURE 6.12
Model of Rev action. Rev is thought to (A) Mediate export of nonspliced or singly spliced RRE-
containing RNAs from the nucleus; (B) Inhibit complete splicing of mRNA; (C) Enhance translation of unspliced and
singly spliced mRNAs. Adapted from Coffin et al. (1997) p. 244.
a mechanism different from that used by Nef. It also facili-
recruits ubiquitin ligases such that hA3G is degraded by the
tates the release of budding virus from the cell, a function
proteasome, thereby preventing it from being incorporated
fulfilled by Env in HIV-2.
into virions.
Vif Abrogates a Cell Defense Mechanism
The dUTPase Gene
Vif, so named because it serves as a viral infectivity fac-
The nonprimate lentiviruses and members of the betaret-
tor, is required for the production of infectious virus in cer-
roviruses contain a gene encoding dUTPase, called dut. This
tain cell types, such as primary CD4 T lymphocytes. These
gene was acquired independently by these two different
cells produce a protein called hA3G or APOBEC3G, which
lineages (Fig. 6.1). The enzyme dephosphorylates dUTP,
deaminates cytosine. This protein is incorporated into prog-
thereby preventing its incorporation into DNA. A cellular
eny virus in the absence of Vif, rendering the virions nonin-
gene usually provides this function for retroviruses (and
fectious because during first-strand DNA synthesis 10% or
for DNA viruses). However, possession of this gene allows
more of cytosines can be deaminated. This results in hyper-
these two genera of retroviruses to replicate in quiescent
mutation of G to A in the plus-sense second-strand DNA,
cells that do not express adequate levels of the enzyme, such
making the provirus nonfunctional. Vif binds to hA3G and
as macrophages.
it is free to diffuse whereas cellular proteins are not, can-
Assembly of Retroviruses
not be excluded. SU/TM is not required for virus assembly.
The retrovirus virion is approximately 100 nm in diam-
Glycoproteins from other viruses can substitute for SU/TM.
eter and acquires its lipid envelope by budding through the
More strikingly, capsids can bud to form noninfectious bald
plasma membrane of the infected cell. Two forms of budding
particles free of glycoprotein. In a key experiment, it has
occur, as illustrated in Fig. 2.21. In the betaretroviruses and
been found that HIV-1 buds from the basolateral surface of
the spumaviruses, the capsid is assembled in the cell cyto-
polarized cells, where SU/TM is found. However, when SU/
plasm and then buds through the plasma membrane. In the
TM is absent, Gag-directed budding occurs from both baso-
alpharetroviruses, the gammaretroviruses, and the lentivi-
lateral and apical surfaces. This result provides evidence that
ruses, the capsid assembles during budding and is not visible
specific interactions between the capsid and the glycopro-
as a distinct structure within the cell. MasonPfizer monkey
teins occur during virus budding and are important, but these
virus, a betaretrovirus, changes from budding of preassem-
interactions are not essential for budding to occur.
bled capsids to assembly of capsids during budding as the
result of a single amino acid change in MA. Thus, the dis-
Alpharetroviruses and Gammaretroviruses
tinction between the two types of budding does not represent
a fundamental difference in budding pathway but simply a
Alpharetroviruses
matter of the stability of the capsid in the absence of interac-
The alpharetroviruses comprise a large collection of
tions with other components of the virion during budding.
avian leukosis and sarcoma viruses. Dozens of ALVs are
The capsid is formed by the assembly of Gag, GagPro, and/
known. They are grouped into seven interference groups,
or GagProPol polyproteins into a structure having spherical
AG, in which members of a group use the same receptor.
symmetry. RNA appears to be required for assembly but not
Infected cells cannot be superinfected by another member
the Env protein, even in viruses whose capsids assemble dur-
of the same interference group, because the receptors for it
ing budding. These polyproteins are incorporated into the cap-
have been eliminated. Expression of the viral envelope pro-
sid in approximately the same ratio as they are produced inside
tein is usually responsible for the downregulation of recep-
the cell. During or after budding, the viral protease cleaves the
tors, which is important for the budding and release of virus.
polyprotein precursors to the final products. These cleavages
It also prevents the cell from becoming infected by hundreds
are required for the virus to be infectious and result in a change
or thousands of progeny viruses. This may be important for
in the structure of the capsid. The final, fully cleaved virion
the survival of the infected cell and may allow it to produce
may have the capsid centrally placed within it or it may be
progeny viruses indefinitely.
eccentrically placed, and the shape of the capsid may be spher-
Interference groups A, B, C, and D of the ALVs are exog-
ical or it may have other shapes, depending on the virus (Fig.
enous viruses that are transmitted as infectious agents from
2.21). As described before, it is important that the cleavages be
chicken to chicken. Members of group E are endogenous
delayed until the virion is partially or completely assembled, or
viruses, resident in the germ line of the chicken. The endog-
the assembly process will not work properly.
enous viruses may be quiescent and not expressed, or parts
During assembly, the genomic RNA is recruited into the
of the provirus genome or even the entire genome may be
‡psid and dimerized. There is a packaging signal in the
RNA, usually referred to as ψ, that is often found in the 5˘
expressed. In the latter case, progeny viruses are formed that
can infect other cells, leading to widespread expression of
region downstream of the LTR. The signal is not present in
the virus in the animal. The endogenous viruses of chick-
the spliced RNAs of many retroviruses but even in those in
ens, as well as those of mammals, tend not to be expressed.
which it is present, the spliced RNAs are not packaged. The
Expression often results in disease and a shortened life span,
packaging signal is not absolutely required for packaging,
and the animals are selected to not express integrated provi-
but increases the efficiency of incorporation into the virion
ruses. In chickens, the level of expression and the time in the
by about 100-fold. Because Gag is the only protein required
animal's life when expression of an endogenous virus occurs
for assembly of capsids, the recognition of the packaging
is different for different strains of chickens.
signal must be a property of Gag. During maturation of the
ALVs are commonly present as infectious agents in
virion, the RNA dimer also matures from a less stable form
chicken flocks around the world. The major illness caused
to a more stable form. tRNA is recruited into the capsid by
by these viruses is a wasting disease characterized by ane-
RT, but association with RT is not absolutely required for
mia, immunosuppression, and poor growth. Perhaps of more
packaging of the primer tRNA.
interest to the molecular biologist, these viruses may also
The envelope glycoproteins, SU and its associated TM,
cause leukemia or sarcoma, as described later.
are incorporated into the virion during budding. The mecha-
Interference groups F and G are viruses of pheasants.
nism by which SU/TM is recruited is uncertain. Evidence
These viruses have not been as well studied as the viruses
indicates that MA interacts with TM, but a model in which
of chickens.
SU/TM is incorporated nonspecifically, perhaps because
Gammaretroviruses
gene under the control of the strong viral promoters, and
the resulting overexpression of the oncogene may result in
The gammaretroviruses consist of a large number of leuke-
a tumor cell. In the case of bursal lymphomas induced by
mia and sarcoma viruses of mice, cats, primates, and other
infection of chickens by ALV, for example, it has been found
mammals. Also included in the genus is reticuloendotheliosis
that more than 80% of tumors have ALV provirus inserted
virus of birds, which causes immunodeficiency. The murine
near the c-myc gene and overexpress the c-myc product. In
leukemia viruses have been particularly well studied. Both
other cases, insertion of the provirus within the oncogene
exogenous viruses and endogenous viruses of mice are known.
may result in the expression of an mRNA that lacks control
The mouse genome contains 5001000 endogenous proviruses
sequences (such as sequences that cause the mRNA to be
that are divided into four classes, but only two of these classes
degraded rapidly) or that is translated into an altered protein
are known to encode infectious viruses. One class encodes
product that has lost regulatory elements. Such a process is
betaretroviruses, described later, and the other class consists of
often seen in erythroblastosis induced by ALV in chickens,
50 to 60 copies of gammaretroviruses. The host range of mouse
for example. Integration of the provirus between two exons
endogenous gammaretroviruses depends on the env gene. These
of c-erbB separates the domain of the protein that binds
viruses may be ecotropic (able to infect only mice), xenotropic
epidermal growth factor and tumor growth factor from the
(unable to infect mice but able to infect other animals, such as
domain that leads to downstream signaling by means of pro-
rats), or polytropic (able to infect both mice and other animals).
tein tyrosine kinase activity. Unregulated signaling by the
The endogenous viruses are usually transcriptionally silent, but
modified protein product results in deregulation of growth.
expression does occur in some mouse strains. In an extreme
A different mechanism of tumor induction is often seen
example, AKR mice usually become viremic at an early age
in mice infected by MLV. Most AKR mice spontaneously
and most of the animals eventually die of leukemia.
express an endogenous virus called AKV1 from birth, and die
Feline leukemia virus (FeLV) is an important pathogen of
of thymic lymphomas in their first year of life. For the tumors
cats. It is an exogenous virus that causes T-cell lymphomas
to develop, multiple recombination events between AKV1
and immunodeficiencies, as well as severe aplastic anemia,
and other endogenous viruses are required to produce an Env
in cats. Sarcoma strains of FeLV are also known. A vaccine
protein with altered host range. The altered SU may stimulate
against FeLV that is given to pet cats is partially effective in
T-cell proliferation by binding the interleukin-2 (IL-2) recep-
preventing FeLV-induced disease.
tor, and other altered interactions with T cells and their pre-
Gammaretroviruses of other mammals, including pri-
cursors may also be important in tumor induction. Alterations
mates, are also known, as are sarcoma viruses derived from
in Env are also found in tumors induced by FeLV.
them. Interestingly, however, no gammaretrovirus is known
It is obvious from this description that the properties of
that infects humans, although the human genome does con-
the virus (host range, the nature of the LTR promoters) as
tain many retroviral-like elements.
well as the properties of the host cell are important in deter-
mining whether a tumor will arise. Thus, different strains
Induction of Leukemia by Alpha-
of the same virus may have different effects upon infection
and Gammaretroviruses
of the same host, or one strain of virus may affect different
strains of its hosts differently.
Alpharetroviruses and gammaretroviruses are important
causes of cancer in chickens, mice, cats, and subhuman pri-
mates. The cancers are usually forms of leukemia or lym-
Alpha- and Gammaretroviruses That
phoma, and arise only after a long latent period. The tumor
Express Oncogenes
cells are usually clonal, having developed from a single pro-
genitor tumor cell. Not all infected animals develop cancer.
Alpha- and gammaretroviruses undergo recombina-
Infection by these retroviruses does not directly produce
tion with cellular oncogenes to produce viruses capable of
tumors. Instead, rare insertional or recombinational events
causing tumors in their hosts. Recombination is thought to
must occur that give rise to a tumor cell. Although these
occur during reverse transcription of a hybrid genome in
events are rare, the very large number of cells infected during
which a host mRNA replaces one copy of the viral RNA.
the persistent infection established by the virus may render
Recombinant viruses that express a variety of oncogenes
such events probable, and after a sufficiently long latent
have been repeatedly isolated from spontaneous tumors in
period the probability that a tumor will arise may be high.
animals that are infected by leukosis/leukemia viruses. Such
It is only recently that the mechanisms by which unmodi-
retroviruses will usually transform cells in culture and rap-
fied alpha- and gammaretroviruses induce tumors have
idly cause tumors when inoculated into a new host. Many of
become at least partially understood. One mechanism
these viruses cause sarcomas and the oncogene-containing
involves the insertion of the provirus near to or within a
virus is then referred to as a sarcoma virus. Examples of
cellular oncogene. This may bring the expression of the
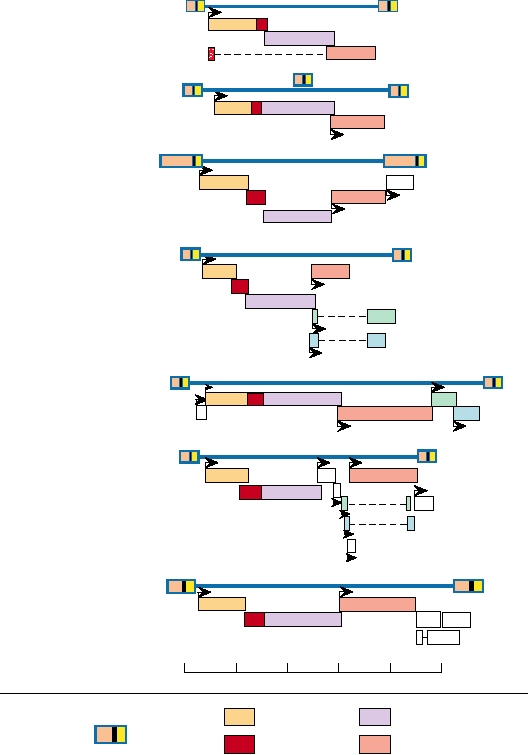
the genomes of four oncogene-containing retroviruses are
shown in Fig. 6.13. Two are derived from avian leukosis
in chickens and transforms cells in culture. The transforming
viruses and two from murine leukemia viruses. These exam-
ability is due to the expression by the virus of the oncogene
ples have been chosen to illustrate a range of possibilities
referred to as src. Many isolates of RSV have been made
for the incorporation and expression of the oncogene in the
over the years, and by definition all such viruses express src.
transforming virus genome.
However, the v-src expressed by the different isolates dif-
Most transforming retroviruses are defective, because the
fer because the recombination points are different, and the
oncogene replaces part of the retrovirus genome. However,
v-src genes usually have mutations that distinguish them
some isolates of Rous sarcoma virus (RSV) are nondefec-
from the cellular gene. Most RSVs are defective because the
tive. In this case, the v-src gene is present in the 3˘ region of
recombination event results in the replacement of parts of
the genome and is translated from an independent, spliced
the viral genome by the cellular oncogene, although nonde-
mRNA. Such a nondefective RSV is illustrated in the figure.
fective RSVs have also been isolated as shown in Fig. 6.13.
Also illustrated is an example of avian myeloblastosis virus,
In the case of defective RSVs, or of other defective onco-
which expresses the v-myb gene. This virus is defective
gene-containing retroviruses, a replication competent ALV,
because the v-myb gene replaces most of the env gene of the
or its equivalent for other retroviruses, is required to sup-
virus. It is translated as a v-myb-env fusion protein from the
ply the missing functions if progeny virions containing the
spliced mRNA that would express the Env protein in ALV.
oncogene are to be produced. However, a defective onco-
The Abelson murine leukemia virus expresses v-abl.
gene-containing virion, once formed, is capable of infecting
This gene replaces the pro and pol genes and part of gag in
a cell, making cDNA, and integrating the proviral DNA into
the example shown, and the virus is defective. In this case,
the host genome without help. Subsequent expression of the
Abl is produced as a fusion protein linked to the N-terminal
oncogene under the control of the viral LTRs leads to trans-
domain of Gag. Moloney murine sarcoma virus expresses v-
formation of the infected cell.
mos. In the example shown, v-mos replaces env and is trans-
Since the discovery of RSV, many different oncogene-
lated from a spliced mRNA normally used to express Env.
expressing retroviruses have been isolated, primarily from
RSV, named for its discoverer Peyton Rous, was one of the
chickens, mice, and cats. The identification and study of
earliest such retroviruses discovered. RSV causes sarcomas
oncogenes has resulted in the discovery of many proteins
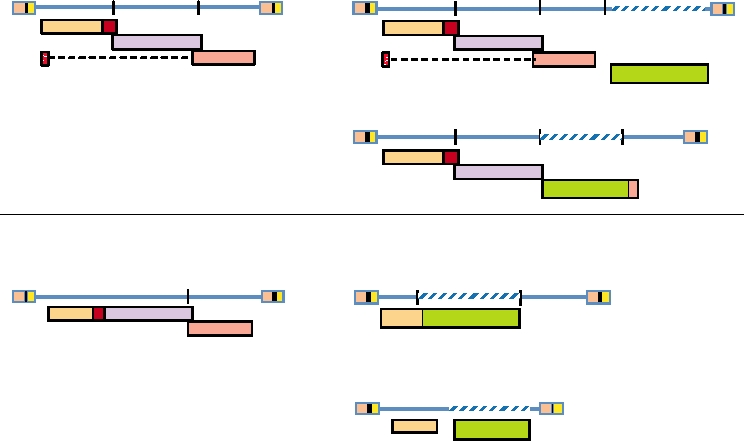
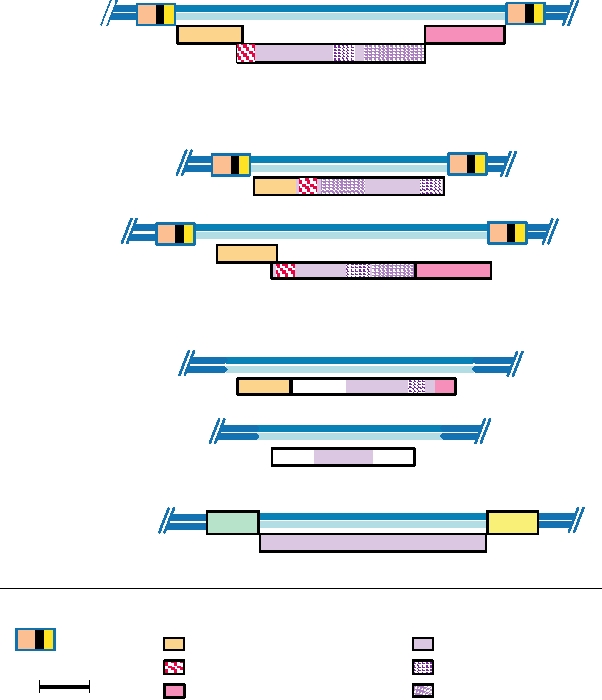
v-onc-containing Retroviral Genome
Retrovirus
Alpharetrovirus
Avian leukosis virus
Rous sarcoma virus (nondefective)
gag-pro
pol
env
src
gag-pro
pol
env
p60 src
Avian myeloblastosis virus (defective)
Ćenv
v-myb
gag-pro
pol
myb-Ćenv
Gammaretrovirus
Murine leukemia virus
Abelson murine leukemia virus (defective)
Ćgag
Ćenv
v-abl
gag-pro-pol
env
Ćgag-abl
Moloney murine sarcoma virus (defective)
gag
v-mos
mos
FIGURE 6.13 Representative v-onc-containing retroviral genomes and the nondefective retroviruses from which they
were derived by capture of cellular oncogenes. In each case the inserted oncogene is shown as a patterned domain in
the DNA and its product as a green box. For Rous sarcoma virus the src gene is expressed from a new spliced message.
For Abelson murine leukemia virus the abl gene is present in a fusion protein with a deleted form of gag. Myb and mos
are expressed from spliced Env messages, from which most or all of the env gene has been replaced by the oncogene.
Adapted from Coffin et al. (1997) pp. 5 and 504 and from Fields et al. (1996), p. 1776.
TABLE 6.5 Selected Retroviral Oncogenes
that are crucial for the regulation of cellular pathways.
A sampling of oncogenes that have been found in retrovi-
Oncogene/
Viral
ruses is given in Table 6.5 to illustrate the range of onco-
functional
onco-
genes that have been captured by retroviruses. Oncogenes
proteina
class
Retrovirus
encode protein kinases, receptors that respond to growth
Growth factors
factors, transcription factors, and other proteins that func-
p28env-sis
tion at critical points in the cell regulatory cycle. The reg-
sis
Simian sarcoma virus
ulation of cell growth is complex. In broad outline, cells
Hormone receptor (thyroid hormone receptor)
express receptors that bind growth factors. On binding its
Avian erythroblastosis virusb
p75gag-erbA
erbA
ligand, the receptor signals the cell to synthesize DNA
Tyrosine kinase growth factor receptors
and divide. This signal is often transmitted by means of
Avian erythroblastosis virusb
gp65erbB
erbB
a phosphorylation cascade by which transcription fac-
gp160env-sea
sea
S13 Avian erythroblastosis virus
tors are activated. The activated transcription factors
gp180gag-fms
fms
McDonough feline sarcoma virus
cause mRNAs to be produced that result in the synthesis
gp80gag-kit
kit
Harvey-Zuckerman-4 feline sarcoma virus
of proteins that, in turn, induce DNA synthesis and cell
p68gag-ros
ros
UR2 avian sarcoma virus
division. These activities are regulated in part by inter-
p68gag-mpl
mpl
Mouse myeloproligerative leukemia virus
actions with regulatory proteins called anti-oncogenes,
gp37eyk
eyk
Avian retrovirus RPL30
‡me of which are described in Chapter 7. Thus, onco-
Nonreceptor tyrosine kinases/ signal transduction factors
genes all have in common that overexpression of the pro-
pp60src
tein, or expression of an altered form of the protein that
src
Rous sarcoma virus
p460gag-abl
no longer responds appropriately to regulatory signals,
abl
Abelson murine leukemia virus
leads to unregulated growth. The regulatory signal could
fpsc
p130gag-fps
Fujinami avian sarcoma virus
be, for example, the ligand to which a growth receptor
fesc
p85gag-fes
Snyder-Theilen feline sarcoma virus
normally responds, or domains that interact with inhibi-
p770gag-actin-
fgr
Gardner-Rasheed feline sarcoma virus
tory elements or that normally require phosphorylation
p90gag-yes
yes
Y73 avian sarcoma virus
for activity, or any one of many other mechanisms that
Serine-threonine kinases/signal transduction factors
regulate cell growth.
p37env-mos
mos
Moloney murine sarcoma virus
For some oncogenes, overexpression by the strong ret-
raf d
p75gag-raf
3611 murine sarcoma virus
rovirus promoter is sufficient for cell transformation. The
mil d
p100gag-mil
MH2 avian myelocytoma virus
large amount of oncoprotein overwhelms the cells' regu-
G Proteins (GTPases)
latory abilities, or the protein is expressed in tissues that
p21ras
H-ras
Harvey murine sarcoma virus
normally do not express it and therefore lack the regula-
p21ras
tory machinery to control it. For most oncogenes, however,
K-ras
Kirsten murine sarcoma virus
overexpression is not sufficient and the viral oncogene,
Transcription factors
referred to as v-onc, differs from its cellular counterpart,
p65gag-jun
jun
Avian sarcoma virus 17
referred to as c-onc. The changes in v-onc render the onco-
p55fos
fos
Finkel-Biskis-Jenkins murine sarcoma virus
protein nonresponsive to regulatory controls so that the cell
p200gag-pol-myc
myc
OK10 avian leukemia virus
is always turned on. One mechanism for the loss of regu-
Avian myeloblastosis virusb
p45myb
myb
latory control is the loss of domains that respond to such
Avian myeloblastosis virusb
p135gag-myb-ets
ets
control signals. A second mechanism is point mutations
p64rel
rel
Avian reticuloendotheliosis virus
that arise during the replication of the recombinant retrovi-
p100gag-maf
maf
Avian retrovirus AS42
rus. Mutations occur at high frequency during the replica-
tion of retroviruses but at a very low frequency during the
a
In these names, p is for protein, gp is for glycoprotein, pp is for
replication of cells.
phosphoprotein, and the numbers are the approximate apparent molecular
The dozens of oncogenes found in retroviruses are too
weight in kD.
numerous to describe here, but a few examples are cited
b
This is a retrovirus with two oncogenes.
c
to illustrate specific points. More than one-third of erythro-
fps and fes are the same oncogene derived from the avian and feline
genomes, respectively.
blastosis in chickens induced by oncogene-containing ALVs
d
raf and mil are the same oncogene derived from the murine and avian
arises from viruses that have captured the erbB gene, and the
genomes, respectively.
viral erbB gene has changes in the C-terminal part of the pro-
Source: Fields et al. (1996) p. 309.
tein (these viruses are called avian erythroblastosis viruses).
that are important in signal transduction. Three different ras
It was noted before that ALV-induced erythroblastosis often
genes have been found in retroviruses and all have changes
results from the provirus integration into the erbB gene.
in codon 12, illustrating the importance of this amino acid
Thus, this gene is very important for the induction of eryth-
for the control pathways that regulate its activity. Mutations
roblastosis. As a second example, Ras proteins are G proteins
in this same codon have also been found in tumors that were
Thus, the probability of transmission of the virus to a new
not produced by retroviruses.
host is low. The appearance of a recombinant retrovirus
Two examples of transcription factor oncogenes will be
expressing an oncogene is an accident that soon disappears
cited. The c-myc gene product is tightly regulated and present
from the population, along with the unfortunate host.
in low amounts in normal cells. It is active on forming het-
erodimers with a second factor, present in larger amounts.
Betaretroviruses
Overexpression of v-myc seems to overwhelm the regulatory
pathways and result in high-level expression of genes required
The best studied member of the betaretroviruses is mouse
for cell proliferation. Even in this case, however, mutations
mammary tumor virus (MMTV). The promoters in the LTR
in v-myc upregulate its transforming ability. Another tran-
respond to estrogen stimulation, consistent with the growth
scription factor, the product of erbA, is a receptor for thyroid
of the virus in mammary epithelium. MMTV occurs as an
hormone. On binding the hormone, it becomes active as a
endogenous virus, present in 04 copies in different strains
transcription factor. v-erbA expresses a modified protein that
of mice. It also occurs as an exogenous virus that infects
no longer needs to bind the hormone in order to be active as
mice by vertical transfer, usually from mother to daughter
a transcription factor, and thus is always on.
through the milk. After a latent period, the virus induces
Often the expression of a single oncogene, whether modi-
mammary tumors with a significant frequency.
fied or not, is not sufficient to achieve full tumorigenic poten-
MMTV encodes a superantigen (the sag gene in Table 6.3).
tial. Some transforming retroviruses have been found to express
As described in Chapter 10, T cells express a receptor that,
two oncogenes, both of which are required for tumor induction.
when stimulated by binding to a peptide antigen recognized
Furthermore, many oncogene-expressing retroviruses induce
by it, causes the cell to become active and divide. Activated
tumors only after a lag and in some cases the tumors are clonal.
T cells induce B cells to proliferate. Superantigens bind to
This suggests that for these viruses, the expression of v-onc is
T-cell receptors and to molecules of the major histocompat-
not in itself sufficient, and additional changes in the infected
ibility complex. Binding is not to the domain of the receptor
cell must occur before it becomes fully tumorigenic.
that interacts with peptide antigen, but to a region shared by
The production of an oncogene-expressing retrovirus that
all T-cell receptors of a given class. A peptide antigen pre-
is capable of inducing a tumor is a rare event, but the presence
sented by major histocompatibility complex molecules may
lead to activation of 10-5 of all T cells. However, the Sag
of the virus is signaled by the tumor itself. Most such viruses
have been isolated from chickens, mice, or cats because these
protein binds to receptors on as many as 10% of all T cells,
animals are commonly infected by C-type retroviruses, but also
leading to their activation. This, in turn, leads to proliferation
because the occurrence of tumors is more likely to be detected
of B cells, which are the first targets of the virus on infec-
in these animals than in many other animals. Chickens are
tion. In this way, the pool of susceptible cells is increased in
processed for human food in very large numbers, and tumors,
size and the infection is amplified. Infected B cells spread the
although rare, are often noticed during processing. Mice are
infection to the mammary glands, where mammary epithe-
used in large numbers in laboratory experiments and receive
lium is infected. The virus induces proliferative changes in
attentive care from investigators. Pet cats are regularly seen
mammary cells, which eventually lead to tumors.
by veterinarians when they become ill.
MasonPfizer monkey virus and the so-called simian
The study of oncogenes that occur in transforming retro-
AIDS virus, or SAIDS virus, are also betaretroviruses. The
viruses has produced a wealth of information about cell reg-
SAIDS virus causes immunodeficiencies in monkeys but is
ulatory pathways by which cell growth and the interactions
distinct from simian immunodeficiency virus (SIV). SIV is
of cells with one another and with various growth hormones
closely related to HIV, described later.
are controlled. Many of these pathways were first discovered
because of the presence of critical components in retrovi-
PTLV Viruses
ruses. Furthermore, some of the oncogenes found in trans-
forming retroviruses were later found to be overexpressed or
The two human T-cell leukemia (or lymphotropic) viruses
to be expressed in a mutant form in human cancers, confirm-
are now usually called primate T-cell lymphotropic viruses
ing the importance of these products in the production of
(PTLV) (Table 6.1). PTLV-1 causes adult T-cell leukemia
human tumors. However, although of great importance for
(ATL) in humans as well as a neurological disorder that has
our understanding of the regulation of eukaryotic cells, and
been termed HTLV-1associated myelopathy (HAM) (from
of importance for the animal in which the tumor produced by
the former name for the virus). The distributions of PTLV-1
the oncogenic retrovirus arose, the transforming retroviruses
and PTLV-2 are shown in Fig. 6.14. PTLV-1 is prevalent
have little significance for the persistence in nature of their
in southern Japan, where transmission appears to be either
retroviral parent. The production of a tumor shortens the life
through sexual intercourse or via breast milk. More than 1
span of the host, and virtually all transforming retroviruses
million people in this region are infected by the virus. The
are defective and require a helper virus for multiplication.
virus is also found in other parts of Asia, in the Caribbean
Equator
Location of PTLV-1 isolates
Areas in which PTLV-1 is endemic
Areas in which PTLV-2 is endemic
Location of PTLV-2 isolates
FIGURE 6.14
Global distribution of PTLV-1 and PTLV-2. Shading indicates the areas of endemism, while the dots
are locations of isolates. For PTLV-1 most of the dots refer to subgroup A isolates, but the B subgroup is found in Zaire,
the D subgroup is found in pygmies in Central Africa, and subgroup C is found in Australia and the islands of Melanesia.
Redrawn from Coffin et al. (1997) Figure 21 on p. 544.
region, and in central Africa, as well as in the United States
encephalopathy in mice, in which inflammation is lacking,
and Western Europe, where it is increasing due to injecting
and many lentiviruses commonly cause neurological disease,
drug use. Blood transfusions have also been implicated in
as will be described later.
the spread of the virus.
PTLV-2 is closely related to PTLV-1. It has been impli-
ATL has a long latent period, 2030 years, and only about
cated as the causative agent of hairy-cell leukemia, but the
1% of infections result in leukemia. Tax is thought to have
number of cases is small and it is not certain that the virus is
a role in induction of leukemia, but the long latent period
in fact responsible for the disease.
implies that other events, probably mutations in the affected
The PTLVs infect not only humans but also a number of
cells, must occur before ATL appears. Patients with acute
nonhuman primates. In any area, the strains of viruses iso-
ATL live only about 6 months on average.
lated from infected humans are more closely related to viruses
HAM is a serious neurological disease. It usually devel-
isolated from monkeys in the region than they are to human
ops more rapidly than ATL, and about 1% of PTLV-infected
strains isolated from another area. Thus, the viruses seem to
humans develop the disease. Demyelination in the spinal
be passed back and forth between monkeys and people.
cord occurs, as well as a vigorous inflammatory response
A virus of cows called bovine leukemia virus (BLV) also
involving T cells and other lymphocytes. It is not known
belongs to this genus of complex retroviruses. This virus causes
whether the pathology is due to damage to virus-infected
B-cell lymphomas in a small fraction of infected animals.
cells by CD8+ T cells (see Chapter 10) or whether the release
of cytokines and chemokines by the inflammatory cells is
Lentiviruses
responsible for the damage.
The retroviruses are best known for their ability to cause
The lentiviruses are complex retroviruses whose core is
tumors, including leukemia, or immunodeficiency, but it is
cone shaped in the mature virion. The most important are
important to note that many retroviruses are associated with
the human immunodeficiency viruses, HIV-1 and -2. Other
neurological diseases. Some of these diseases are character-
members of the genus include a number of simian immu-
ized by an inflammatory response, as is HAM, whereas in
nodeficiency viruses (SIV), feline immunodeficiency virus
others there is no inflammation associated with the disease.
(FIV), bovine immunodeficiency virus (BIV), visna virus
As examples, many strains of MLV cause a spongiform
(which causes disease in sheep), equine infectious anemia
virus (EIAV), and caprine arthritis-encephalitis virus (CAEV)
The primary cellular targets of HIV in infected humans
are macrophages and CD4+ T cells. These cells express CD4,
(Table 6.1). As described earlier, the regulation of lentivirus
replication is complex because lentiviruses encode three to
the primary receptor for all primate lentiviruses, as well as
six accessory genes, depending on the virus, to regulate the
chemokine coreceptors that are also required for entry of the
replication cycle and aid in the assembly of progeny viruses.
virus (Chapter 1). As described in Chapter 1, strains of HIV
All lentiviruses have a specific tropism for macrophages,
are known that preferentially infect macrophages (M-tropic
which comprise the major reservoir of infected cells in an
virus) or that preferentially infect T cells (T-tropic), which
animal. Other cells can also be infected, and this can be
is a function of the env gene of the virus. The infection of
important in the disease process (e.g., CD4+ T cells in HIV
macrophages and of CD4+ T cells differs in several impor-
infection). Lentiviruses establish a lifelong, chronic infec-
tant respects. HIV can replicate in fully differentiated, non-
tion which elicits a vigorous immune response that is unable
dividing macrophages, but infection of macrophages does
to clear the infection. Most known lentiviruses cause seri-
not lead to extensive cytopathology. Efficient replication of
the virus in CD4+ T cells, in contrast, requires that the T cell
ous disease in their native host after a long latent period.
However, SIV produces an asymptomatic, lifelong infec-
be activated (Chapter 10) and results in the death of the cell.
Most infections of CD4+ T cells do result in the death of the
tion of its natural host, African monkeys. Intriguingly, SIV
causes AIDS when transmitted to Asian monkeys.
cell, but the virus is also able to establish a latent infection
in resting memory T cells, which has important implications
for antiviral therapy as described later.
Human Immunodeficiency Virus
A representative time course of the infection of a human
The two human lentiviruses, HIV-1 and HIV-2, cause
by HIV-1 is shown in Fig. 6.15. Primary infection may be
the well-known disease called AIDS (acquired immunode-
asymptomatic (most infected people do not see a doctor on
ficiency syndrome). Although both HIV-1 and HIV-2 cause
primary infection) or accompanied by nonspecific symp-
AIDS in people, HIV-2 is not as serious a pathogen. The
toms that appear 36 weeks after infection. Symptoms may
latent period before disease develops is longer for HIV-2,
include rash, fever, diarrhea, arthralgia, myalgia, nausea,
the virus is not as easily transmissible, and it has not spread
sore throat, lethargy, headache, or stiff neck. The number
of CD4+ T cells declines during this phase. An immune
as extensively as has HIV-1. It is HIV-1 that is responsible
response is mounted that includes both CD8+ cytotoxic T
for the vast majority of AIDS in people, and HIV-1 has cor-
respondingly been much more exhaustively studied.
cells (CTLs) and antibody production (Chapter 10). This
Primary
Death
Infection
1200
Acute HIV Infection
107
1000
Opportunistic
Anti-HIV Immune
Infections
Responses
106
800
Clinical Latency
105
Onset of
600
Symptoms
104
400
AIDS
103
200
102
0
3
6 9 12
2
4
6
8
10
12
Weeks
Years
Time after infection
FIGURE 6.15 Typical time course of HIV infection and progression to disease. Patterns of CD4+ T-cell decline and
virus load vary greatly among patients. See also Fig. 6.16 for mortality as a function of initial viral load. Some case
definitions for AIDS specify a CD4+ count below 400 cells/ml (horizontal line). Adapted from Coffin et al. (1997) Figure
5 on p. 600.
In individuals in which viral replication is high, the CD4+
reduces the amount of virus in the blood, the number of
CD4+ T cells recovers but does not reach preinfection levels,
T-cell count declines faster and the time to appearance of
and symptoms of disease ameliorate. However, clearance
AIDS is shorter. The rate of virus replication is probably a
of the virus is not complete, and a long period of clinical
function of the strength of the immune response against the
virus, especially the CD8+ CTL response. Individuals with
latency ensues. During this period, even though the infected
a better immune response control virus replication better
individual is often asymptomatic, the virus continues to rep-
and longer. Once the symptoms of AIDS appear, the time
licate actively, especially in lymph nodes, and the immune
to death is usually 12 years unless antiviral treatment is
system continues to fight the infection. It has been estimated
that during this clinically quiescent period 108 to 1010 virus
administered.
CD4+ T cells, most of which function as T helper cells, are
particles per day are released into the peripheral blood sup-
ply and cleared. During this time the turnover of CD4+ lym-
a critical part of the immune system. They are necessary for
phocytes is also 108 to 109 cells per day.
an immune response to an antigen, whether the response is
to produce CD8+ CTLs or to produce circulating antibodies
In untreated individuals, the continuing replication of
virus results in a steady decline in the number of CD4+ T
(see Chapter 10). AIDS results when so few CD4+ T cells are
cells (Fig. 6.15) and the continuing appearance of mutations
present that the body is unable to mount an effective immune
in the virus. In the Env protein, the rate of change in the
response. A partial list of symptoms that appear and oppor-
amino acid sequence averages about 2.5% per year, with a
tunistic infections that occur is shown in Table 6.6. The first
symptoms usually occur when the CD4+ T-cell concentration
large fraction of changes occurring in certain hypervariable
falls below 500/µl of blood and may include reactivation of
regions. It seems clear that this accumulation of mutations is
driven, at least in part, by immune selection.
viruses such as herpes zoster, reactivation of bacteria such as
Mycobacteria tuberculosis, oral lesions caused by fungi, or
lymph node pathology. These first symptoms are sometimes
AIDS, the Disease
referred to as AIDS-related complex or ARC. Full-blown
AIDS is normally signaled by a drop in the CD4+ T-cell con-
With time the immune system is overwhelmed, with
centration below 200/µl. At this point the infected individual
the result that the replication of HIV escalates, opportu-
nistic fungal, protozoal, bacterial, and viral infections occur,
becomes susceptible to numerous opportunistic infections,
characteristic cancers appear, and neurological symptoms
including those of protozoa such as Pneumocystis carinii,
become apparent. The time to progression to AIDS follow-
bacteria such as Mycobacterium tuberculosis, and fungi such
ing infection by HIV is variable but averages about 10 years.
as Candida albicans. Viruses that are normally controlled
Symptoms occur much sooner in infants infected at birth,
by the immune system become a problem, such as cytome-
perhaps because the immune system is not as well devel-
galovirus and herpes simplex virus. Several virus-induced
oped. In adults infected by the virus, the time to appearance
cancers become common, such as lymphoma caused by
of the symptoms of AIDS is correlated with the level of viral
EpsteinBarr virus, Kaposi's sarcoma caused by human her-
replication during the latent period, as illustrated in Fig. 6.16.
pesvirus 8, or anogenital carcinoma caused by human papilloma
CD4+cells/m l
Viral RNA copies/ml
100
100
>800
≤4500
500-800
300-500
75
75
4500-13000
<300
50
50
13000-36000
25
25
>36000
0
0
0
2
4
6
8
10
0
2
4
6
8
10
Time (years)
Time (years)
FIGURE 6.16 Relationship between the extent of HIV replication or the CD4+ cell count (two different measures of
HIV/AIDS) and clinical progression of disease and time to death. KaplanMeier curves are shown for survival versus
viral load (left) and concentration of CD4+ T cells (right) at time of diagnosis, plotted by quartiles. Redrawn from Mellors
et al. (1996).
TABLE 6.6
Pathology and Opportunistic Infections of Patients with HIV/AIDS
Type of
Stage of HIV/AIDS
Syndrome/symptoms
infectious agent
Organism
CD4+ T-Cell count 200500 cells/ml
Generalized lymphadenopathy
Headache, fever, malaise
Generalized weight loss
Shingles
Viral
Reactivation of herpes zoster
Skin lesions
Viral
Molluscum contagiosum
Basal cell carcinomas of skin
Oral lesions
Thrush
Fungal
Candida albicans
Hairy leukoplakia
Viral
EpsteinBarr virus
Lung disease (tuberculosis)
Bacterial
Reactivation of Mycobacterium tuberculosis
CD4+ T-cell count <200 cells/ml
(Syndromes in addition to those mentioned above)
Microbial Infections
Pneumonia
Protozoan
Pneumocystis carinii
Disseminated toxoplasma
Protozoan
Toxoplasma gondii
Severe diarrhea
Protozoan
Isospora belli
Chronic diarrhea
Protozoan
Cryptosporidia
Tuberculosis
Bacteria
Mycobacterium tuberculosis
Bacterial infections
Bacteria
Salmonella, Streptococcus, Hemophilus,
Cryptococcus
CNS disease
Fungal
Cryptococcus neoformans
Viral Infections and
PML
Viral
JC polyomavirus
Malignancies
Disseminated disease of lungs, brain, etc.
Viral
Genital herpes, cytomegalovirus
B-cell lymphoma
Viral
EpsteinBarr virus, HHV-8
Kaposi's sarcoma
Viral
HHV-8
Anogenital carcinoma
Viral
Human papilloma virus
Cervical carcinoma (in women)
Viral
Human papilloma virus
Other Syndromes
Wasting disease
??
??
Aseptic meningitis
AIDS dementia complex
Viral
HIV encephalopathy
Abbreviations: CNS, central nervous system; PML, progressive multifocal leukoencephalopathy; HHV-8, human herpesvirus eight.
Source: Adapted from Coffin et al. (1997) Table 1, p. 597.
this terminal phase, when the immune system can no longer
virus. Many of these opportunistic diseases, such as P. cari-
control viral infection (Fig. 6.15). Early loss of gut-associated
nii pneumonia or Kaposi's sarcoma, are rarely seen in non-
lymphoid tissue (GALT) is an effect of HIV infection and a
HIV-infected people. Others are regularly seen in the general
harbinger of poor prognosis.
population but HIV-infected individuals have a much higher
The lymph nodes are organs that trap invading patho-
incidence of the disease, 100-fold higher in the case of lym-
gens and present them to immune cells. Large numbers of
phoma, for example. Other symptoms of AIDS include a
macrophages and CD4+ T cells are present in lymph nodes,
wasting syndrome and neurological abnormalities, described
and the nodes are sites of active HIV replication. During
later. The symptoms of AIDS become progressively worse
as the CD4+ count continues to drop until virtually no CD4+
the progression of disease following HIV infection, the
cells are present. The decline in numbers of CD4+ cells is
lymph nodes deteriorate. During the final stages of disease,
the architecture of the nodes is completely destroyed and
accelerated by the increased replication of HIV itself during
this loss of lymph node function contributes to the loss of
At the end of 2005, an estimated 40 million people were
immune function.
living with HIV/AIDS, with about 1 million of those in
A wasting syndrome is characteristic of late-stage HIV
North America (Table 6.7). The cumulative death toll from
infection. Bowel involvement results in diarrhea and malab-
AIDS since the beginning of the epidemic now exceeds 25
sorption. The wasting syndrome is thought to be a symptom
million. Five million new infections occurred worldwide in
of HIV infection itself, but opportunistic infections of the
2005, and 3 million people died of AIDS that year. In the
gut probably exacerbate the symptoms in many individuals.
absence of treatment, virtually all infected people go on to
Most HIV-infected individuals suffer, sooner or later,
develop AIDS and die within 2 years of the appearance of
from neurological disease. Some disease is caused by oppor-
the symptoms of AIDS.
tunistic pathogens, such as progressive multifocal leukoen-
The focus of HIV infection is sub-Saharan Africa, where
cephalopathy caused by JC virus (Chapter 7). However,
25 million people, or more than 7% of the population, are
two-thirds of infected individuals develop an encephalopa-
thought to be infected and where medical care is still primi-
thy induced by HIV infection itself that produces symptoms
tive. In the course of the epidemic from 1984 to 2005, the
including dementia, motor and behavioral abnormalities, and
number of HIV infections increased dramatically (Fig. 6.17;
seizures. These symptoms are referred to as AIDS dementia
see also Table 6.7). The extent of the problem is illustrated
complex. HIV is present in the brains of infected individuals
by the fact that in some large cities in sub-Saharan Africa,
in macrophages and microglia, but does not infect neurons.
an estimated 2040% of adults may be infected. The spread
The cause of the neuronal loss induced by HIV infection is
of HIV has resulted in a marked decrease in life expectancy,
unknown.
which has reversed a long period of increasing life expect-
ancy as health care standards have improved (Fig. 6.18).
In other areas of the world, HIV continues to spread but
Spread of AIDS
has not reached the extreme levels found in parts of Africa
HIV is spread sexually, through contaminated blood, and
(Table 6.7). In the Americas and in Southern and Southeast
from mother to child. The virus is present in semen, both
Asia, about 0.6% of the population is infected. Figure 6.19
as free virus and in infected cells, and in vaginal secretions
shows a breakdown of the incidence of AIDS by state in
of infected people, and can be transmitted by either homo-
the United States in 2003. Fig. 6.20 shows the explosive
sexual or heterosexual intercourse. The probability of a
increase in AIDS prevalence in Asia between 1991 and
woman becoming infected during unprotected vaginal inter-
2001. In Western Europe and Australia, estimates are that
course with an infected male is estimated at less than 1/50.
0.3% of the population is infected. Table 6.7 documents the
The risk of a man becoming infected during heterosexual
spread of the virus between 1996 and 2005.
intercourse is less. The risk is much higher if genital lesions
In North America, HIV has been spread primarily
resulting from sexually transmitted diseases are present and
through homosexual contacts and by sharing of needles by
in the absence of circumcision. The risk of infection is also
drug users who inject drugs, but the frequency of hetero-
much higher during receptive anal intercourse. The use of
sexual transmission is increasing. In developing countries,
condoms reduces the risk of transmission by a large factor.
heterosexual contact is the primary mode of transmission.
The virus can also be spread by means of contaminated
The result is that in Africa, the male/female infection ratio
blood, whether through blood transfusions, the use of con-
is approximately 1 to 1, whereas in the United States the
taminated products by hemophiliacs, by needle stick in
male/female ratio is almost 4 to 1 because of the importance
health care workers, the sharing of needles by injecting drug
of male homosexual contacts in the spread of the virus in
users, or via tattoo needles. Blood transfusion was an impor-
this country (Table 6.7). In the United States, the number of
tant source of infection before the development of tests for
newly diagnosed cases of AIDS, especially in white homo-
the virus and remains a problem in developing countries
sexual men, has dropped because awareness of the disease
where blood tests may not be regularly available. Similarly,
has led to the more frequent use of condoms and a lowered
many hemophiliacs became infected before the development
incidence of multiple sex partners, and because more effec-
of blood tests. Use of blood tests and screening of donors
tive drug therapies have been introduced (Fig. 6.21). This
for risk factors have greatly reduced the risk of transmis-
trend may be changing for the worse, however, as the devel-
sion of HIV through blood products in developed countries.
opment of more effective drugs to treat the disease appears
However, transmission among drug users remains an impor-
to have resulted in an increase in unprotected sexual activity.
tant source of infection.
Even as the death rate has declined, the total number of per-
Untreated, infected women transmit the virus to a new-
sons living with HIV/AIDS continues relentlessly upwards.
born child about one-third of the time. Transmission can
The capability of HIV to spread rapidly is illustrated by
occur both during delivery and during breast-feeding. The
its recent spread in Thailand. In Thailand, every male is
use of anti-HIV drugs has reduced transmission to infants in
subjected to compulsory military service and is tested for
countries where the drugs are available.
HIV infection at the age of 18 when called up for military
TABLE 6.7 Characteristics of the Global HIV/AIDS Epidemic
HIV/AIDS in 2005
Number of people with
HIV/AIDS
Deaths from HIV/AIDS
Adult
Primary
Geographical
% Change
% Change
prevalence
mode of
region
1996
2005
19962005
1996
2005
19962005
%Women
rate (%)
transmission
Comments 2005
North America
750,000
1.2 million
60
61,300
18,000
-71
26
0.7
1. MSM
New diagnoses
2. IDU and
predominantly in
hetero
African Americans
Caribbean
270,000
300,000
11
14,500
24,000
65
53
2.3
Hetero
Haiti has highest
prevalence at 5.6%
Latin America
1.3 million
1.8 million
38
70,900
66,000
-7
27
0.6
1. MSM
Brazil accounts for
2. IDU and
>33% of the infected
hetero
persons in this region
Sub-Saharan
14 million
25.8 million
84
783,700
2.4 million
206
51
7.2
Hetero
Sub-Saharan Africa has just
Africa
over 10% of the world
population and 60% of
people living with HIV
North Africa and
200,000
510,000
155
10,800
58,000
437
37
0.2
1. IDU
80% of infected persons in
Middle East
2. Hetero
this region are in Sudan
W. Europe
510,000
720,000
41
21,000
12,000
-43
28
0.3
1. MSM
Most infected persons have
2. IDU and
access to treatment
hetero
C. and E. Europe
50,000
1.6 million
3100
1,000
62,000
6100
26
0.9
1. IDU
Most in the Russian
and Central Asia
2. MSM
Federation, and in
persons <30 years old
South and Southeast 5.2 million
7.4 million
42
144,000
480,000
233
30
0.6
Hetero
Epidemic now fueled by new
Asia
infections in India
East Asia
100,000
870,000
770
1,200
41,000
3320
22
0.1
1. IDU and
Even low prevalence translates
hetero
into large numbers of
2. MSM
infected persons, primarily
in China
Oceaniaa
13,000
74,000
469
1,000
3600
260
20
0.2
1. MSM
In New Zealand, 85% of new
2. IDU and
transmission is in MSM
hetero
Abbreviations: IDU, injecting drug use; hetero, heterosexual transmission; MSM, sexual transmission among men who have sex with men.
Percent change was calculated as [(Figure for 2005/Figure for 1996) 1] × 100.
Areas with unusually high prevalence or percent increases are highlighted in red, and reduced numbers are shown in blue.
a
Includes Papua New Guinea, which has the highest prevalence in the region, and Australia and New Zealand which have two of the lowest.
Source: Data through the end of 2004 is from the website: www.unaids.org/epidemic_update/report/ and updated to 2005 with information from "UNAIDS/WHO AIDS Epidemic Update:
December 2005." At the end of 2005 there were 40.3 million people living with HIV/AIDS; during 2005 there were 4.9 million new infections, 3.1 million deaths, 43% of the infected were women,
and the overall prevalence rate was 0.63%.
1991
1984
2001
1999
2004
Percentage of Adults
(15 - 49 yrs)
Infected with HIV
20.01-36%
10.01-20%
5.01-10%
1.01-5%
0-1%
no data
FIGURE 6.17 Explosion of the prevalence of infection with HIV in Africa during the last 20 years. The panels show
the percentage of adults (15 to 49 years old) infected with HIV in various countries of Africa in 1984, 1991, 1999, 2001,
and in 2004. This map was redrawn from Nations Programmes on HIV/AIDS Conference in Geneva, June, 2000 reported
in Schwartlander et al. (2000), and updated with information from the 2006 Report on the Global AIDS Epidemic, from
the UNAIDS Programme.
duty. Twenty years ago HIV was almost unknown in
widely, with the result that the rate of new infections has
Thailand, but within a very few years the disease became
dropped dramatically (Fig. 6.22).
prevalent, infecting some 3% of the male population.
Thailand is a country that is very tolerant of sexual activity
Prevention and Treatment of AIDS
and many young males visit prostitutes. Most of the pros-
titutes became infected with HIV and began to transfer the
The most successful method for the prevention of
infection to many of their male partners, resulting in the
most viral diseases has been the development of vaccines.
observed explosion of infections in the population. In some
However, the development of a vaccine against HIV has
provinces in northern Thailand, as many as 20% of 18-year-
been a very slow, frustrating, and to date unsuccessful proc-
old males were infected with HIV as of 15 years ago. The
ess. This is perhaps not surprising in light of the fact that the
government began an aggressive education campaign about
virus establishes a persistent lifelong infection in the face
the disease and how it is acquired and distributed condoms
of a vigorous immune response on the part of the host that
65
Botswana
60
55
Zimbabwe
50
Zambia
Uganda
45
40
Malawi
35
30
1960
1970
1980
1990
2000
Year
FIGURE 6.18 Changes in life expectancy in selected African countries with high HIV prevalence, 1955 to 2005.
Adapted from UNAIDS publications slide 12 of the AIDS in Africa series on the Web site www.UNAIDS.org. and
updated information from the UN Human Development Report 2004. The points plotted at 2005 are projections.
16
527
52
2
7
37
179
242
757
6722
184
25
13
102
676
8
733
1906
1514
75
60
216
775
1734 506
75
95
279
1572
786
368
220
111
404
5967
1102
835
214
778
Rate:
189
628
111
Cases/100,000
1907
509 471
population
3413
1048
<5
4744
5 to 9.9
110
10 to 14.9
District of Columbia
Puerto Rico
≥15
17
961
1065
Virgin Islands
34
FIGURE 6.19 AIDS in the United States in 2003. In 2003, a total of 44,232 cases of AIDS were reported in the United
States. In this map, the rate (cases per 100,000 population) is indicated by the color, and the total number of reported cases
is given for each state. The rate for the District of Columbia is so far off scale (169) that a special color is used. Data from
MMWR, Summary of Notifiable Diseases 2003.
1991
Prevalence of HIV infection in
Adults 15 to 49 years old
2.01% - 5%
1.01% - 2%
0.51% - 1.0%
0.1% - 0.5%
<0.01%
No Data Available
Outside Reporting Region
2001
FIGURE 6.20 Prevalence of HIV/AIDS in Asia in 1991 and 10 years later, in 2001, in Adults aged 15 to 49 years.
Note that even at a prevalence of less than 2%, due to its enormous population, India now has roughly 6 million persons
living with HIV/AIDS. Adapted from 2006 Report on the Global AIDS Epidemic, from the UNAIDS Programme.
succeeds in controlling the virus for many years. Vaccine
of behavior, especially sexual behavior. Safe sex practices,
development continues to be a priority, but to the current
such as the use of condoms, have lowered the numbers of
time only public education to slow the spread of the disease
new infections, especially for HIV infections among gay
and the development of antiviral drugs have had any success
white men. Education campaigns in Thailand and Uganda
in control of the virus.
have succeeded in slowing the spread of the disease, and
Much effort has been made to reduce the epidemic spread
similar campaigns in other areas of the world have met with
of HIV by educating the public and thereby altering patterns
at least some success. However, although such campaigns
AIDS cases
New AIDS case definition
450
90
Deaths
400
Persons living with AIDS
80
350
70
Monotherapy
(primarily nucleoside inhibitors)
300
60
250
50
200
40
150
30
100
20
Multidrug therapies with
50
10
protease inhibitors
0
0
93
85
86
87
88
89
90
91
92
94
95
96
97
98
99
00
01
02
03
Year of diagnosis or death
FIGURE 6.21 Estimated number of AIDS cases and deaths per year of diagnosis among persons >13 years old in the
United States from 1985 to 2003 and number of Americans living with AIDS. Data from AIDS Surveillance-Trends at:
http://www.cdc.gov/hiv/graphics/images/.
160
140
120
100
80
60
40
20
0
1985
1987
1989
1991
1993
1995
1997
1999
2001
2003
FIGURE 6.22
New HIV infections per year in Thailand from 1985 to 2003. From: Thailand's Response to HIV/AIDS:
Progress and Challenges (2004) United Nations Development Programme report. Figure 2 on p. 2.
slow the spread of the virus, they do not stop it and will
these analogues into DNA results in premature chain ter-
never succeed in eliminating the virus from the population.
mination, and because the viral RT has a high affinity for
The development of drug therapies for control of HIV
these drugs whereas cellular enzymes have a low affinity for
infection has met with at least partial success. Early drugs for
them, their primary effect is to disrupt replication of HIV.
treatment of HIV infection were disappointing at best (see
However, virus mutants that are resistant to the drugs rapidly
Fig. 6.21). The drug 5˘ azidothymidine (AZT) and related
appear, and control by this agent was found to be of lim-
nucleoside analogues were first used in monotherapy, and
ited duration. Second-generation inhibitors were developed
were initially successful in decreasing virus load and allow-
that interfere with the activity of the viral protease, which is
ing partial recovery of immune function. Incorporation of
required for assembly of infectious virus, or with the viral
integrase. The protease inhibitors have been widely used, and
T cells, although other sources of persistent virus may be
variants resistant to them appear much more slowly, proba-
present. In any event, it appears that the therapy will have to
bly because multiple changes in the protease are required for
be continued over the lifetime of the patient.
resistance. However, resistant virus does eventually emerge
Although HAART has been successful in many patients,
when the inhibitor is used in monotherapy.
it is clearly not a panacea. First, it is very expensive and
Third-generation drug therapy, called highly active
suitable for use only in developed countries that can afford
antiretroviral therapy or HAART, uses a combination of two
a high level of health care. Second, there is significant tox-
different nucleoside analogues and a protease inhibitor. This
icity that results from the drug treatment, making lifelong
treatment has been quite successful in more than half of HIV
treatment problematic. Third, compliance becomes a major
patients, in which the viral load declines to undetectable lev-
issue when any therapy, especially therapy with significant
els with full recovery of immune function. The introduction
side effects, must be continued indefinitely. It may be for
of combined therapy has led to a marked reduction of the
such reasons that the therapy fails in a considerable fraction
death rate from AIDS in the United States (Fig. 6.21). Figure
of HIV patients. Fourth, resistant variants arise, necessitat-
6.23 illustrates that by 1995 AIDS had become the leading
ing changes in the regime of drugs being used. However,
cause of death in U.S. adults 2544 years of age. With the
although this treatment is complex and of limited success,
advent of HAART, AIDS has fallen to the level of homicide
its very existence seemingly has led to a rise in unprotected
as a killer of young adults.
sexual activity and to an upsurge in the rate of HIV infection
At first it was hoped that long-term treatment with this
among populations at risk.
regime would result in the elimination of the virus and cur-
Attempts to develop a vaccine continue and there is hope
ing of the infection. However, this does not appear to be pos-
that a vaccine may ultimately be produced. It is clear that
sible. Virus production quickly resumes when HAART is
an immune response that involves both neutralizing anti-
bodies and CD8+ T cells is important in the control of the
discontinued, even in patients that have been in treatment
virus during the clinically latent state. The T-cell response
for many years without detectable virus production dur-
seems to be especially important. Stronger CTL responses
ing that time. Reemergence of the virus may occur because
result in lower levels of virus production during the latent
the treatment does not eliminate the virus latently infecting
30
Injuries
25
HIV-AIDS
20
Cancer
Unintentional Injuries
Heart Disease
15
Cancer
Heart Disease
Homicide
Suicide
Suicide
Liver Disease
10
Stroke
Diabetes
Homicide
Pneumonia and Influenza
HIV and AIDS
5
0
1980
1985
1990
1995
2000
2005
Year
FIGURE 6.23 Annual number of deaths in the United States in adults 2544 years old for each of the 10 leading causes
of death, normalized for population growth. The data shown are the aggregate for all races and both sexes, for the years 1980
to 2003. Data came from CDC, National Center for Injury Prevention and Control and National Vital Statistics System.
state, which results in a slower rate of progression to AIDS.
human viruses and humans are the only reservoir. This topic
Neutralizing antibodies are also important, but it has been
is covered in more detail in Chapter 8 on Emerging Viral
found that fresh isolates of virus from patients are difficult
Diseases.
to neutralize, presumably because of the many carbohydrate
chains attached to the HIV surface glycoprotein and because
Nonhuman Lentiviruses
of the unusual folding properties of this protein. In addition,
variants arise during prolonged infection that are resistant to
SIV, FIV, and BIV cause AIDS or an AIDS-like disease in
the mix of antibodies that exist in the patient at the time that
animals. At least 36 different primate species in sub-Saharan
the variants arise.
Africa are infected by different SIVs. These viruses cause no
In developing a vaccine, there is the significant question
disease in their natural hosts but do cause AIDS in Asian mon-
as to whether sterilizing immunity, which is very difficult
keys. The disease in Asian monkeys appears very similar to
to achieve, is required, or whether protection from disease
human AIDS, and SIV has been useful in laboratory studies as
can be obtained with nonsterilizing immunity. For virtually
a surrogate for human AIDS. The primary receptor for SIV is
all other viruses for which vaccines have been developed,
CD4, as is the case for HIV. FIV causes immunodeficiency in
the vaccine is nonsterilizing--subsequent infection with the
cats that appears to result by mechanisms similar to those that
virulent virus results in a subclinical infection that is quickly
produce human AIDS, although the primary receptor for the
virus is different. The number of CD4+ T cells declines mark-
damped out. But with a virus that produces a lifelong persis-
tent infection in the face of an immune response, the needs
edly in the late stages of the disease and the disease is charac-
may be different. Recent findings with a very small group
terized by opportunistic infections as well as by neurological
of patients have suggested that nonsterilizing immunity
symptoms. BIV infection of cattle results in persistent kill-
may suffice, at least in some cases, but more information is
ing of lymphocytes and lesions in the central nervous system,
needed before the complete answer is known.
symptoms that resemble those produced by HIV.
A second, related question is whether is it possible to
In contrast, equine infectious anemia virus (EIAV) in horses,
boost the immune response in people infected with HIV and
caprine arthritis-encephalitis virus (CAEV) in goats, and visna
obtain better control or elimination of the virus. Because of
virus in sheep produce quite different symptoms. EIAV causes
the very long latent period before disease develops, this is
recurrent anemia in horses and is an important veterinary prob-
an extremely difficult question to approach. Various clini-
lem. Primary infection by EIAV results in an acute disease that
cal trials have been conducted to test such possibilities, a
is soon controlled by an immune response. However, variants
topic that is covered in Chapter 11. Clinical trials have also
arise that are resistant to preexisting antibody and cause recur-
been conducted that test the responses of uninfected people
rences of fever and acute anemia. This continues for 6 months
to immunization with the external glycoprotein of HIV. To
to 1 year, after which the infection becomes clinically inappar-
date, such trials have been disappointing.
ent. Infection is lifelong, however, as in all lentiviruses. It is
The possibility of a live virus vaccine is suggested by the
thought that the virus binds to, but does not infect, red blood
finding that a small fraction of infected people exists who
cells. These cells are then destroyed by a complement pathway
have not progressed to AIDS after many years of infection
or by engulfment by macrophages, resulting in the anemia.
and whose CD4+ T-cell count has remained fairly stable. In
The virus also appears to suppress the differentiation of precur-
at least some of these individuals, the infecting HIV strain
sors to red blood cells. Although anemia is the characteristic
has deletions in nef. Initially, experiments with nef deletions
disease produced by EIAV, in a small number of horses virus
in SIV in monkeys give comparable results--the monkeys
infection results in encephalomyelitis.
did not develop AIDS and they were protected for some time
CAEV infection of goats produces an inflammatory
from infection by wild-type strains of SIV. However, they
response that often results in arthritis. The arthritis arises
eventually did succumb to AIDS, illustrating the difficulties
several years after infection and resembles rheumatoid
in developing a live virus vaccine for a virus that establishes
arthritis in humans. Other symptoms produced by infec-
a persistent infection that is ultimately fatal, and for which
tion include encephalomyelitis in 20% of infected animals,
the symptoms of disease are delayed for many years.
which arises within 6 months of infection. Visna virus infec-
tion of sheep follows a similar course, but the characteristic
disease produced is pneumonia, which arises several years
The Origins of HIV
after infection. Encephalomyelitis occurs in less than 5% of
Like measles virus, HIV is a zoonotic disease. HIV-
infected animals.
1 derives from SIV that infects chimpanzees and HIV-2
derives from SIV that infects sooty mangabey monkeys.
Spumaviruses
HIV-1 has entered the human population and become estab-
lished at least three times in the last century, and HIV-2
The spumaviruses comprise a genus of complex retro-
became established independently. These viruses are now
viruses. There are no known human spumaviruses. A virus
and prokaryotic chromosomes have been able to evolve.
previously isolated from human cells in culture, and referred
The number of different kinds of retroelements is conse-
to as human foamy virus or human spumaretrovirus, is now
quently very large and encompasses not only the retrovi-
thought to be a virus of chimpanzees or other monkeys. The
ruses described before but also endogenous retroviruses,
spumaviruses group with epsilon- and gammaretroviruses
retrotransposons of various types, retrointrons, retroplas-
(Fig. 6.1), but differ in many properties from other retrovi-
mids, and retrons, among others. A partial listing of these
ruses. The Gag polyprotein is not cleaved to produce MA,
elements is given in Table 6.8, and the phylogenetic rela-
CA, and NC proteins (Fig. 6.7). Spumaviruses appear to bud
tionships among the RTs of those elements that possess RT
into the endoplasmic reticulum, rather than from the plasma
are shown in the tree in Fig. 6.24.
membrane as do other retroviruses. In addition, Pol is trans-
As a class, retroelements are found in all organisms from
lated from its own mRNA, rather than from the genomic
prokaryotes to mammals. Many of these elements are capa-
RNA as a fusion protein with Gag (Fig. 6.6). Recent studies
ble of amplification and are mobile, able to move within
have also indicated that the RNA of human spumavirus is
the genome. Some encode RT, whereas others utilize RT of
converted to DNA during packaging of the virion. It is pos-
other elements for their propagation. These elements can be
sible that virions may contain either DNA or RNA, since
thought of as selfish DNA. They replicate to fill up the host
both are found in virus preparations. The packaging of DNA
genome to what is, in essence, its carrying capacity. These
in the virion rather than RNA, the translation of Pol from its
elements must be benign and have only limited effects on
own mRNA, and budding into the endoplasmic reticulum,
the replication potential of the organisms in which they
resemble the events that occur in the pararetroviruses such
reside. Otherwise their host would be selected against dur-
as the hepadnaviruses, described later. In other respects, the
ing evolution. The number of retroelements in eukaryotic
spumaviruses resemble retroviruses rather than pararetro-
genomes is often very large. In mammals, 510% of the
viruses. They use a tRNA primer for reverse transcription,
genome appears to consist of endogeneous retroviruses,
like the retroviruses, and their DNA integrates into the host
and all retroelements together comprise about half of the
genome as a provirus. Thus, they are retroviruses that are
genome. The endogeneous retroviruses are provirus-like,
intermediate in their properties between the classical retrovi-
containing LTRs and primer binding sites flanking regions
ruses and the pararetroviruses.
with detectable relationships to gag and pol. Some of these
are active endogenous viruses that have inserted into the
Retroelements in the Genomes of
Living Organisms
TABLE 6.8 Retroelements and Their Distribution in Nature
Reverse transcriptase is an ancient enzyme. Its origins
probably go back to the RNA world at the time of the inven-
Class
Distribution
tion of DNA, when such an enzyme would have been useful
Eukaryotic retroelements
for converting informational RNA molecules into DNA, the
form in which genetic information is now stored. It appears
Retroviruses (Retroviridae
Vertebrates
Table 6.1)
to resemble the RNA polymerases of RNA viruses more than
Pararetroviruses
DNA polymerases or DNA-dependent RNA polymerases,
and may have evolved from a primordial RNA polymerase
Hepadnaviruses (Hepadnaviridae, Mammals, birds
Table 6.10)
of the RNA world. The enzyme still exists in modern eukary-
Caulimoviruses
Plants
otes and plays a role in the replication of eukaryotic cells.
Telomerase is a eukaryotic enzyme that repairs the ends of
Retrotransposons
LTR retrotransposonsa
the linear chromosomes, which are progressively shortened
Animals, plants, fungi, protozoa
during replication. Cells lacking telomerase cease to divide
Non-LTR Retrotransposons
Animals, plants, fungi, protozoa
after the ends of the chromosomes become too short to sup-
Mitochondrial elements
port replication. Telomerase is a ribonucleoprotein enzyme
Group II introns (retrointrons)
Mitochondria of fungi and plants,
that synthesizes repeat sequence elements that are attached
plastids of algae
to the end of the chromosomes. These elements are specific
Mauriceville plasmid
Mitochondria of Neurospora
for a given species. This process involves reverse transcrip-
RTL gene
Mitochondria of Chlamydomonas
tion of an RNA template that is a component of the telomer-
Prokaryotic retroelements
ase, and the ends of the chromosomes serve as primers. The
msDNA-associated RTb
Myxococcus xanthus, E. coli, other
composition of telomerase has not been completely eluci-
bacteria
dated, but it does have a reverse transcriptase activity that is
related to the RT present in a variety of retroelements.
a
Includes the two families Pseudoviridae and Metaviridae; see Table 6.9.
There has been a long period of time in which elements
b
msDNA, multicopy single-stranded DNA.
that utilize reverse transcription for insertion into eukaryotic
Source: Adapted from Eickbush (1994), Table 1, p. 122.
Name
Host
Family
Genus
Other Groupings
DHBV
Vert.
Orthohepadnavirus
HBV
Hepadnaviridae
WHBV
Pararetroviruses
Caulimovirus
Plants
CoYMV
Caulimoviridae
CaMV
Plants
Badnavirus
D. m.
412
Metavirus
D. m.
Ty3
Metaviridae
D. m.
Gypsy
Errantivirus
D. m.
17.6
Plants
Ted
MMTV
Betaretrovirus
MPMV
Alpharetrovirus
RSV
Retroviridae
Retroviruses
Vert.
Deltaretrovirus
PTLV
Lentivirus
HIV
Gammaretrovirus
MLV
Spumavirus
HSRV
Copia
D. m.
Hemivirus
D. m.
1731
Pseudoviridae
Tst1
Plants
Pseudovirus
Ty1
Yeast
Yeast
Telomerases
Human
L1
Human
R2
D. m.
I Factor
D. m.
Poly A transposons
Tad
Neurospora
D. m.
Jockey
D. m.
R1
al1
Yeast
Retrointrons
al2
Mito
Neurospora
Retroplasmids
Ec67
Bacteria
Retrons
Mx65
RNA dependent RNA polymerases
FIGURE 6.24 Phylogenetic tree of the retroelements, based upon the sequences of the reverse transcriptases. The blue
boxes enclose the retroviruses and pararetroviruses. Plant hosts are shown in green, insects in brown, fungi in purple,
vertebrates in blue, and humans in red. The vertical distances are arbitrary; the length of the horizontal branches is
proportional to the number of changes. The tree has been rooted with the RNA-dependent RNA polymerase sequences.
Abbreviations of the retroviruses were given in Table 6.1; abbreviations of orthohepadnaviruses are found in Table 6.10.
Names of representative Pseudoviridae and Metaviridae are given in Table 6.9; the Sirevirus and Semotivirus genera, as
well as four more genera of Caulimoviridae, which are exclusively plant viruses, are not shown. CoYMV is commelina
yellow mottle and CaMV is cauliflower mosaic virus; D. m. is Drosophila melanogaster (fruit fly). Redrawn from Coffin
et al. (1997) Figure 17 on p. 411, plus information from Fauquet et al. (2005), p. 418.
genome recently, whereas others are inert. The time at
ALVs into the chicken genome occurred after chicken and
which any element entered the germ line can often be esti-
turkey separated.
mated from comparative studies of these elements in dif-
Next, four classes of eukaryotic elements, the endog-
ferent animals, and these proviruses or other retroelements
enous retroviruses, the LTR-containing retrotransposons,
constitute a kind of fossil record for these elements. As one
the poly(A)-containing retrotransposons, and the group II
example, chickens have one to four endogenous proviruses
retrointrons, are described in more detail. The relationships
closely related to the ALVs, but there are no such endog-
among these elements and their relationships to the infectious
enous viruses in turkey or quail. Thus, insertion of these
retroviruses described earlier, and to the pararetroviruses
described later, are illustrated by the phylogenetic tree in
although in some animals one or more endogenous retro-
Fig. 6.24. The genetic organizations of these elements are
viruses are normally expressed during the lifetime of an
compared in Fig. 6.25.
animal.
The best studied endogenous retroviruses are those of
chickens and of mice. Mice contain more than 1000 endog-
Endogenous Retroviruses
enous viruses or elements that are thought to have originated
Endogenous retroviruses are proviruses that have become
from endogenous viruses. Most of these are defective and
established in the germ line of many different organisms at
unable to undergo a complete cycle to give rise to infectious
various times in the past. They align with the retroviruses in
virus, but up to 100 copies of nondefective endogenous ret-
the tree in Fig. 6.24, and their genetic organization is identi-
roviruses are present in some strains of mice. As described
cal to that of the simple retroviruses (see Fig. 6.25). Most are
before, these viruses can be ecotropic, xenotropic, poly-
defective, having deletions or mutations in them that prevent
tropic, or modified polytropic, depending on the receptor
them from undergoing a full round of replication. Some are
recognized by their envelope gene. In most strains of mice,
not defective, however, and can, upon activation, give rise to
these viruses are not expressed, although in many cases
progeny virus that can infect other cells. Most endogenous
they can be induced to replicate by treatment of the cell
retroviruses are silent--the genes that they encode are not
with mutagenic agents or agents that lead to demethylation
expressed or are expressed only under restricted conditions,
of DNA. In some strains of mice, the endogenous viruses
Retroviridae
LTR
LTR
Pol
Gag
Env
LTR Retrotransposons
Pseudoviridae
LTR
LTR
Copia
ORF
LTR
LTR
Metaviridae
Gypsy
ORF 2
ORF 1
ORF 3
Non-LTR Retrotransposons
(TAA)n
I Factor
(Drosophila)
ORF 1
ORF 2
AAAAA
R2
(Drosophila)
RT
ORF
Group II Intron
Exon 1
Exon 2
RT
ORF
Polymerase motifs
LTR
Gag-related sequences
Reverse transcriptase
Protease
RNase H
1 kb
Integrase
Env-related sequences
FIGURE 6.25
Comparative genome organizations of retroviruses, retrotransposons, and retrons. In each case the
integrated form of the element is shown, flanked with dark blue bars representing the host DNA. ORFs for each element
are shown below the DNA, and boxes on different lines represent different reading frames. Note that Gypsy has now been
classified as an Errantivirus in the family Metaviridae, and Copia is a Hemivirus in the family Pseudoviridae. Various
motifs related to retroviruses are indicated with different colors and shadings, as shown in the key. At the 3˘ end of many
non-LTR retrotransposons there are a variable number of TAA repeats (I factor) or adenylate residues (R2). Adapted from
Eickbush (1994).
are transcriptionally active and expressed at some stage dur-
retrotransposons are obviously related to retroviruses. The
ing the lifetime of the mouse without having to be induced.
LTRs that they possess are closely related to retroviral LTRs,
Early expression and vigorous replication in strains of mice
including the conserved two terminal nucleotides of the pro-
such as AKR leads to the development of leukemia in most
virus. They have tRNA primer binding sites and polypurine
of these mice.
tracts, so that reverse transcription of the RNA appears to
An interesting question deals with how the xenotropic
follow the same pathways as in retroviruses.
viruses, which cannot infect mice, entered the mouse germ
Thus, the LTR-containing retrotransposons are effec-
line. The simplest explanation would be that these viruses
tively intracellular viruses that replicate only within an
entered the germ line of an ancestor of the modern mouse
individual cell. As such they have recently been classified
that did express receptors for the virus. In this model, dur-
into two families of virus-like agents, the Pseudoviridae
ing the evolution of mice these receptors were silenced or
and the Metaviridae (Table 6.9). These agents are ancient,
mutated to a form that was no longer usable by the xeno-
being found in plants, insects, and fungi as well as in animals
tropic viruses. It is possible that modern mice were selected
including vertebrates. The argument has been made that the
to not express a functional receptor for these viruses, in order
Metaviridae gave rise to the current retroviruses, that they
to minimize the virus load from endogenous viruses.
represent an ancestral form of retroviruses in which an RT-
containing element became mobile by associating with gag-
like genes that allowed it to spread within the cell. Later
LTR-Containing Retrotransposons
acquisition of an envelope gene in vertebrate Metaviridae
The LTR-containing retrotransposons are essentially
gave rise to a complete retrovirus. The Metaviridae lineage
endogenous retroviruses that lack the envelope gene (Fig.
is a sister lineage to the Retroviridae as shown in Fig. 6.24,
6.25B). They contain LTRs and encode Gag-related pro-
consistent with this hypothesis.
teins, protease, and RT with its associated activities. RNA
The two families of LTR retrotransposons, which rep-
transcribed from the elements is translated to produce Gag
resent two independent evolutionary lines (Fig. 6.24), each
and Pol, and the RNA is assembled into intracellular cap-
contain three genera (Table 6.9). In the Metaviridae are Ty3
sid-like particles that reverse transcribe it back into DNA.
of yeast and Gypsy of Drosophila, among other elements.
Subsequent integration into the genome allows the elements
They are closely related to the pararetroviruses of plants
to spread within the genome. Further, the LTR-containing
(Fig. 6.24). The Pseudoviridae contain Ty1 of yeast and
TABLE 6.9 LTR Retrotransposons
Family/genera/
Type virus name
type virus
abbreviation
Transmission
Distribution
Comments
Pseudoviridae (LTR retrotransposons of the Ty1-copia family)
Pseudovirus
Exclusively vertical
Worldwide
Genus contains several other viruses of
Saccharomyces cerevisiae
SceTy1V
plants and fungi
Ty1 virus
Hemivirus
Unknown
Worldwide
Found in insect, fungal, and viral genomes
Drosophila melanogaster
DmeCopV
copia virus
Serivirus
Unknown
This genus found only in plants
Glycine max SIRE1 virus
GmaSIRV
Metaviridae (LTR retrotransposons of the Ty3-gypsy family)
Metavirus
Unknown
Genus contains several other viruses of
plants, insects, and fungi
Saccharomyces cerevisiae
SceTy3V
Ty3 virus
Errantivirus
Unknown
Most infect insects
Drosophila melanogaster
DmeGypV
gypsy virusa
Semotivirusa
Unknown
Primarily in nematodes, but also insects
and vertebrates
Ascaris lumbricoides
AluTasV
Tas virus
a
All members of these genera have a env-like ORFs, but the ORFs of errantiviruses have no sequence similarity to those of semotiviruses.
Source: Fauquet et al. (2005).
Search WWH :