Minus-Strand RNA Viruses
INTRODUCTION
OVERVIEW OF THE MINUS-STRAND
RNA VIRUSES
Seven families of viruses contain minus-strand RNA
[(-)RNA], also called negative-strand RNA, as their
Viruses belonging to four families of (-)RNA viruses, the
genome. These are listed in Table 4.1. Included in the table
Paramyxoviridae, the Rhabdoviridae, the Filoviridae, and
are the names of the genera belonging to these families
the Bornaviridae, contain a nonsegmented RNA genome
and the hosts infected by these viruses. Six of the fami-
having similar organization. They are grouped into the order
lies are known to contain members that cause epidemics
Mononegavirales (mono because the genome is in one piece,
of serious human illness. Diseases caused by these viruses
nega for negative-strand RNA). This was the first order to
include influenza (Orthomyxoviridae), mumps and mea-
be recognized by the International Committee on Taxonomy
sles (Paramyxoviridae), rabies (Rhabdoviridae), encepha-
of Viruses and still is one of only three orders currently rec-
litis (several members of the Bunyaviridae), upper and
ognized. Viruses belonging to the other three families, the
lower respiratory tract disease (numerous viruses in the
Arenaviridae, Bunyaviridae, and Orthomyxoviridae, pos-
Paramyxoviridae), and hemorrhagic fever (many viruses
sess segmented genomes with two, three, and six to eight
belonging to the Bunyaviridae, the Arenaviridae, and the
segments, respectively. Regardless of whether the genome
Filoviridae), as well as other diseases. Bornavirus, the sole
is one RNA molecule or is segmented, the genomes of all
representative of the Bornaviridae, also infects humans
(-)RNA viruses possess a similar suite of genes, as illus-
and may cause neurological illness, but proof of causality
trated in Fig. 4.1. In the Mononegavirales, the order of genes
is lacking. Many of the (-)RNA viruses presently infect
along the genome is conserved among the viruses (although
virtually the entire human population at some point in time
the number of genes may differ). In the viruses with seg-
(e.g., respiratory syncytial virus, influenza virus), whereas
mented genomes, the genes can be ordered in the same way
others did so before the introduction of vaccines against
if the segments are aligned as shown. In addition, many
them (e.g., measles virus and mumps virus). These viruses
features of virion structure and of replication pathways are
are thus responsible for a very large number of cases of
shared among the (-)RNA viruses.
human illness. The diseases caused by such widespread
viruses are usually serious but have a low (although not
Structure of the Virions
insignificant) fatality rate. In contrast, some (-)RNA
viruses, such as rabies and Ebola viruses, cause illnesses
All (-)RNA viruses are enveloped and have helical
with high fatality rates but (fortunately) infect only a small
nucleocapsids. The different families encode either one
fraction of the human population. The (-)RNA viruses
or two glycoproteins (called G in most of the families but
are major causes of human suffering, and all seven fami-
called HA, NA, F, or HN in some, after hemagglutinating,
lies and the viruses that belong to these families will be
neuraminidase, or fusion properties). These glycoproteins
described here.
are present in the viral envelope. In most cases, cleavages
TABLE 4.1 Negative-strand RNA Viruses
Type virusa
Host(s)b
Family/genus
Genome size (in kb)
Transmission
Mononegavirales (nonsegmented)
Rhabdoviridae
1316
Vesiculovirus
VSIV
Vertebrates
Some arthropod-borne
Lyssavirus
Rabies
Vertebrates
Contact with saliva
Ephemerovirus
BEFV
Cattle
Arthropod-borne
Novirhabdovirus
IHNV
Fish
Two genera of plant viruses
Arthropod-borne
Filoviridae
13
Marburgvirus
Marburg
Vertebrates
?
Ebolavirus
Zaire Ebola
Vertebrates
?
Paramyxoviridae
1620
Respirovirus
Sendai
Vertebrates
Airborne
Morbillivirus
Measles
Vertebrates
Airborne
Rubulavirus
Mumps
Vertebrates
Airborne
Henipavirus
Hendra
Vertebrates
Airborne
Avulavirus
Newcastle disease
Birds
Airborne
Pneumovirus
HRSV
Vertebrates
Airborne
Metapneumovirus
TRTV
Turkeys
Airborne
Bornaviridae
Bornavirus
~9
BDV
Vertebrates
Contaminated forage
Segmented Negative Strand RNA Viruses
Orthomyxoviridae
1014.6
Influenzavirus A
Influenza A
Vertebrates
Airborne
8 segments
Influenzavirus B
Influenza B
Vertebrates
Airborne
Influenzavirus C
7 segments
Influenza C
Vertebrates
Airborne
Thogotovirus
6 segments
Thogoto
Vertebrates
Arthropod-borne
Isavirus
8 segments
ISAV
Fish
Waterborne
Bunyaviridae
1120 in 3 segments
Orthobunyavirus
Bunyamwera
Vertebrates
Mosquito-borne
Hantavirus
Hantaan
Vertebrates
Fecesurinesaliva
Nairovirus
Dugbe
Vertebrates
Tickborne
Phlebovirus
Rift Valley fever
Vertebrates
Arthopod-borne
Tospovirus
TSWV
Plants
Thrips
Arenaviridae
1014 in 2 segments
Arenavirus
LCMV
Vertebrates
Urinesaliva
a
Abbreviations of virus names: VSIV, vesicular stomatitis Indiana virus; BEFV, bovine ephemeral fever virus; IHNV, infectious hematopoietic necrosis
virus; HRSV, human respiratory syncytial virus; TRTV, turkey rhinotracheitis virus; BDV, Borna disease virus; ISAV, infectious salmon anemia virus;
TSWV, tomato spotted wilt virus; LCMV, lymphocytic choriomeningitis virus.
b
In all cases, "Vertebrates" includes humans.
are required to produce the mature glycoproteins, such as
All (-)RNA viruses have a single major nucleocapsid
cleavage to release a signal peptide, cleavage to separate two
protein (called N) that encapsidates the virion RNA to form
glycoproteins produced as a common precursor, or cleav-
the helical nucleocapsid. Also present in the nucleocapsid
age to activate viral infectivity. The glycoproteins project
is a phosphorylated protein that is required for RNA syn-
from the lipid bilayer as spikes that are visible in the electron
thesis, variously called P (for phosphoprotein) or NS (for
microscope (see, e.g., Fig. 2.18D).
nonstructural protein because it was not originally known to
MONONEGAVIRALES
39
59
RHABDOVIRIDAE
VESICULOVIRUS
N
P/C
L
M
G
tr
le
(VSIV)
PARAMYXOVIRIDAE
N
SH
L
P/V
F
HN
M
RUBULAVIRUS
tr
le
(SV5)
FILOVIRIDAE
p30
FILOVIRUS
M1
N
P
sGP
GP
L
M2
tr
le
(VP35)
(VP40)
(VP30)
(VP24)
(ZEBOV)
BORNAVIRIDAE
N
P/X
M
G
L
BORNAVIRUS
le
tr
(BDV)
SEGMENTED NEGAVIRALES
BUNYAVIRIDAE
S RNA
M RNA
L RNA
BUNYAVIRUS
N NSs
G1
L
G2
NSm
(SSHV)
S RNA
L RNA
ARENAVIRIDAE
ARENAVIRUS
L
N
G2
G1
NS
(LCMV)
ORTHOMYXOVIRIDAE
RNA 3 INFLUENZAVIRUS A
RNA 1
RNA5
RNA8
RNA4
RNA6
RNA 2
(FLUAV)
PB1
PA
PB2
HA
NA
N
NS1/NS2
M1/M2
Nucleocapsid Genes
Glycoprotein Gene(s)
Polymerase Gene(s)
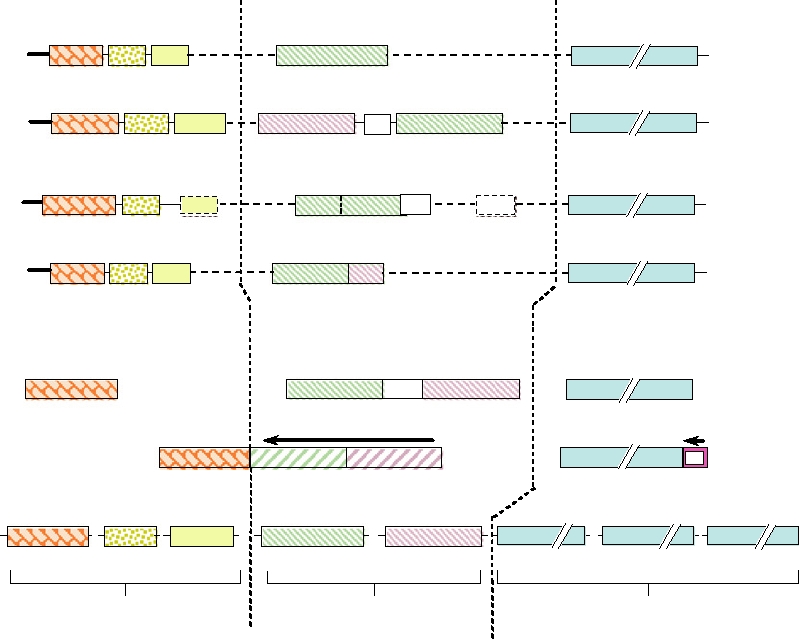
FIGURE 4.1 Genome organizations of the Negavirales. The genomes of representatives of the four families of
Mononegavirales have been aligned to illustrate functional similarity between gene products. The individual gene segments
of the representatives of the three families with segmented genomes, Bunyaviridae, Arenaviridae, and Orthomyxoviridae,
have been aligned according to similarity of function with those of the Mononegavirales above. Gene expression strategies
for the other genera of Bunyaviridae vary (see Fig. 4.21). Abbreviations of virus names are as follows: VSIV, vesicular
stomatitis Indiana virus; SV5, simian virus 5; ZEBOV, Zaire ebolavirus; BDV, Borna disease virus; SSHV, snowshoe
hare virus; LCMV, lymphocytic choriomeningitis virus; FLUAV, influenza A virus. The gene products are abbreviated as
follows: le is a leader sequence; N is the nucleoprotein; P is the phosphoprotein; M (M1, M2) are matrix proteins; G (G1,
G2) are membrane glycoproteins; F is the fusion glycoprotein; HN is the hemagglutinin-neuraminidase glycoprotein;
L is the RNA polymerase; NA is the neuraminidase glycoprotein; HA is the hemagglutinin glycoprotein; NS (NV, SH,
NSs, NSm) are nonstructural proteins; PB1, PB2, and PA are components of the influenza RNA polymerase; tr is the
trailer sequence. Within a given genome, the genes are drawn approximately to scale. mRNAs for most genes would be
synthesized left to right; however, an arrow over a gene means that it is in the opposite orientation (ambisense genes).
Redrawn from Strauss et al. (1996), Figure 5.
be a component of the virion), as well as a few molecules of
spherical in the electron microscope. The example of influ-
an RNA-dependent RNA polymerase. The polymerase is a
enza virus is shown in Figs. 2.1 and 2.22D, and the paramyxo-
large, multifunctional protein called L in most families but
virus measles virus is shown in Fig. 2.22C. The compositions
is present as three proteins in the Orthomyxoviridae. L and P
of these virions are not rigorously fixed and some variability
form a core polymerase that replicates the viral genome and
in the ratios of the different components, particularly in the
synthesizes mRNAs.
glycoprotein content, is present. The rhabdoviruses are bullet
A matrix protein (M) is present in all of the viruses
shaped or bacilliform and appear more regular (Fig. 2.23), but
except the bunyaviruses and the arenaviruses. M underlies
even here variations in the composition of the glycoproteins in
the lipid bilayer where it interacts with the nucleocapsid. M
the envelope can occur. The filoviruses are filamentous (Fig.
also inhibits host transcription and shuts down viral RNA
2.23). Orthomyxoviruses and paramyxoviruses also produce
synthesis prior to packaging.
filamentous forms as well as round virions (see Fig. 2.25E). In
The (-)RNA virions are heterogeneous to a greater or
fact, clinical isolates of influenza viruses and human respira-
lesser extent. Members of five families often appear roughly
tory syncytial virus are predominantly filamentous.
In contrast to the translation strategy used by the (+)RNA
Synthesis of mRNAs
viruses, the (-)RNA viruses do not produce polypro-
For all (-)RNA viruses, the first event in infection is the
teins that require processing by virally encoded enzymes,
synthesis of mRNAs from the minus-strand genome by the
and virus-encoded proteases are unknown among them.
RNA polymerase present in the nucleocapsid. Because this
However, most of the glycoproteins of the (-)RNA viruses
polymerase is necessary for the production of the mRNAs,
are produced as precursors that are processed by cellular
and because the proteins translated from the mRNAs are
enzymes, and some of these precursors can be considered
required for replication of the genome, the naked genomes
to be polyproteins.
of (-)RNA viruses are not infectious, nor are complemen-
tary RNA copies of the genomes. It has been possible, none-
theless, to rescue virus from cDNA clones of viral genomes
Replication of the Genome
by using special tricks, as described in Chapter 11.
Multiple mRNAs are produced from minus-strand
Replication of the (-)RNA genome requires the pro-
genomes. By definition, each region of the genome from
duction of a complementary copy of the genome, called
which an independent mRNA is synthesized is called a
an antigenome or virus-complementary RNA (vcRNA),
gene. In (-)RNA viruses with segmented genomes, it is
which is distinct from the mRNAs (schematically illus-
obvious that multiple mRNAs are produced (the number
trated in Figs. 1.11C and D). Neither the genomic (-)RNA
of mRNAs produced actually exceeds the number of seg-
nor the antigenomic template produced during replication
ments, as described later). In the Mononegavirales, multiple
is ever free in the cytoplasm. Instead, replication of the
mRNAs arise from the use of a single polymerase entry site
genome, as well as the synthesis of mRNAs, takes place in
at the 3′ end of the genome. The polymerase then recog-
nucleocapsids (sometimes referred to as ribonucleoprotein
nizes conserved start and stop signals at the beginning and
or RNP), which always contain the phosphoprotein and the
end of each gene to generate discrete mRNAs. The amount
polymerase as well as N and the viral RNA. Replication
of mRNA produced for any given gene is controlled by the
can only occur in the presence of ongoing protein synthe-
location of the gene relative to the single polymerase entry
sis to produce the new proteins required to encapsidate the
site, because mRNA synthesis is obligatorily sequential and
genome or antigenome. The mRNAs can be synthesized in
attenuation occurs at each gene junction. Thus, more mRNA
the absence of viral protein synthesis and lack encapsida-
for the proteins encoded 3′ in the genome is made and more
tion signals, so that they are released into the cytoplasm
protein is thus translated from these genes. The N protein,
where they can associate with ribosomes and be translated.
required for encapsidation of both genome and antigenome,
Thus, early after infection, mRNAs are synthesized. After
is thereby produced in the largest quantities and the RNA
translation of the mRNAs, which leads to production of
polymerase, needed in the smallest quantities, is made in the
sufficient amounts of viral proteins, a switch to the produc-
smallest quantities. The synthesis of mRNAs is described in
tion of antigenomes for use as templates occurs, followed
more detail in the sections on Rhabdoviridae.
by production of genomic RNA from the antigenomic
Most of the mRNAs are translated into a single protein,
templates.
but a few of the genes produce mRNAs that are translated
The genomes (or genome segments) of all (-)RNA
into more than one product. Multiple products can be pro-
viruses have sequences at the ends that are complementary
duced from the same gene by the use of alternative trans-
(so-called inverted terminal repeats). In the bunyaviruses,
lation initiation codons during translation of an mRNA; by
the RNAs form panhandles, circular structures that are vis-
the introduction of nontemplated nucleotides during mRNA
ible in the electron microscope. Panhandles have also been
synthesis, which results in a shift in the reading frame; or by
reported for influenza A virus. In other viruses, circles have
splicing of an mRNA. The P genes, in particular, of most
not been seen but may form transiently during replication.
of the (-)RNA viruses are translated into multiple products,
It is possible that these complementary sequences exist to
and two of the segments of influenza virus, which replicates
promote cyclization of the RNA, which may be required
in the nucleus, can be spliced to produce a second mRNA
for replication of the genome or synthesis of mRNAs. It has
encoding a different product. In no case are the mRNAs exact
been shown for influenza A virus that the viral RNA repli-
complements of virion RNAs. This is obvious in the case
case interacts with both ends of the RNA during synthesis of
of the Mononegavirales, where as many as 710 mRNAs
RNA, similar to the story for alphaviruses and flaviviruses
are produced from a single long genomic RNA, but is also
described in Chapter 3. Another possible explanation for the
complementary sequences is that the promoter at the 3′ end
true of the segmented (-)RNA viruses, where the mRNAs
lack cis-active sequences required for encapsidation and
of the genomic RNA that is recognized by the viral RNA
replication that are present near the ends of the antigenome
synthetase for the production of antigenomes is the same, at
least in part, as the promoter at the 3′ end of the antigenomic
segments. Thus, the mRNAs of (-)RNA viruses do not rep-
licate nor are they packaged into virions.
RNA that is used to initiate the production of genomic RNA.
Search WWH :