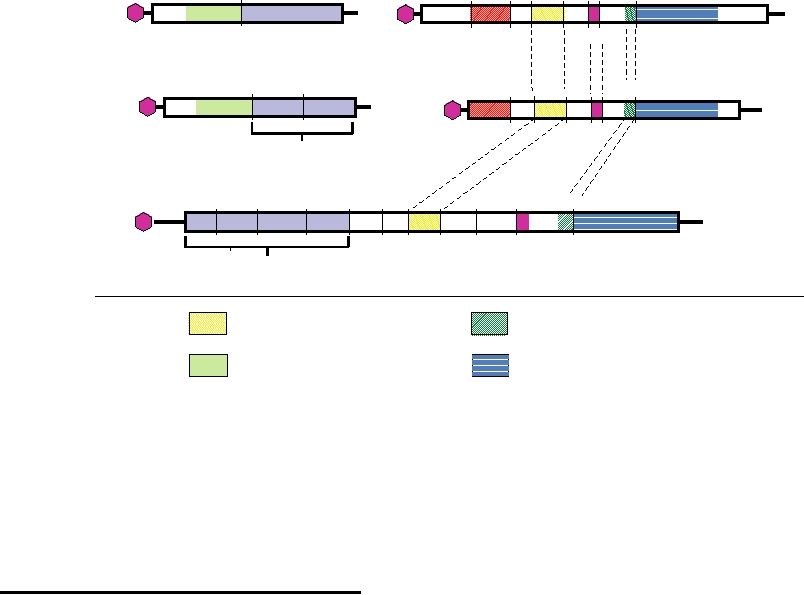
RNA2
RNA 1
NEPOVIRUS (TBRV)
32k
58k
25k
An
An
VPg
VPg
Polymerase
Coat Protein
VPg
MP
COMOVIRUS (CPMV)
24kpro
32k
58k
An
An
VPg
MP
Polymerase
VPg
VPg
Structural Proteins
POLIOVIRUS
3Cpro
An
2Apro
VPg
VPg
Polymerase
2C
Structural Proteins
Replication Complex
Cysteine Protease
Helicase
Movement Protein
Polymerase
FIGURE 3.42 Comparison of the genomes of bipartite como- and nepoviruses and monopartite poliovirus. Domains
in the helicase, polymerase, and protease that share sequence homology over long stretches of amino acids are identified
with differently colored patterns. The related 32k proteins of como- and nepoviruses and the movement proteins encoded in
RNA 2 (MP) have no counterpart in poliovirus. TBRV, tomato black ring virus; CPMV, cowpea mosaic virus. The structural
proteins of the three viruses show no sequence similarity. Adapted from Strauss and Strauss (1997) Figure 2.12.
be distinguished that probably diverged from one another
ORIGIN AND EVOLUTION OF PLUS-STRAND
early in the evolution of RNA viruses. Most RNA viruses
RNA VIRUSES
also possess an RNA helicase that is required to unwind the
RNA during replication. These helicases also appear to have
A reasonable hypothesis for the origin of the RNA
diverged from a single source, but three lineages can be dis-
viruses is that they began as an mRNA that encoded an RNA
tinguished here as well. A third shared function in those RNA
polymerase. The acquisition of an origin of replication that
viruses with capped mRNAs is a methyltransferase gene (an
allowed the mRNA itself to be replicated by its encoded
activity required for capping), and two methyltransferase
product would give rise to a self-replicating RNA and could
lineages can be distinguished. Finally there are the viral pro-
have represented the first step in the development of a virus.
teases that process polyproteins. The two distinct types of
Subsequent recombination with an mRNA encoding an
proteases with independent origins are the proteases derived
RNA-binding protein that could be modeled into a capsid
from serine proteases (which may possess serine or cysteine
would give rise to a very simple virus. This protovirus could
at the active site) and the papain-like proteases. The different
then evolve through continued mutation and recombination
lineages of these four core activities have been reassorted in
into something more complex. In support of this idea is the
various ways during the evolution of the RNA viruses, as
fact that the capsid protein of a large number of viruses,
shown in the figure.
including bacterial viruses, plant viruses, and animal viruses
The second mechanism for divergence among viruses is
that are otherwise unrelated to one another, share a common
mutation. Lack of proofreading in RNA replication means
fold, suggesting that once a proper capsid protein arose it
that the mistake frequency during replication is very high, on
was retained during the evolution of many viruses while
the order of 10-4. Most mistakes are deleterious and do not
being modeled into new shapes.
persist in the population. However, because the mistake fre-
Examples of the importance of recombination in the
quency is so high, many different sequences can be tried rap-
evolution of RNA viruses have been discussed. Computer-
idly because of the rapid replication rate of viruses. The net
aided studies that have attempted to align the amino acid
result is that the rate of sequence divergence in RNA viruses
sequences of the proteins of different (+)RNA viruses have
is very high, up to 106-fold faster than their eukaryotic hosts.
suggested that all these viruses share core functions that have
Three studies of the rate of sequence divergence in RNA
common ancestral origins. These results are summarized in
viruses are illustrated in Fig. 3.44. In these studies, regions
Fig. 3.43. All RNA viruses possess an RNA polymerase and
of the genomes of viruses isolated over a period of many
these all appear to have derived from a common ancestral
years were compared. The rates of sequence divergence in a
source. However, three lineages of RNA polymerases can
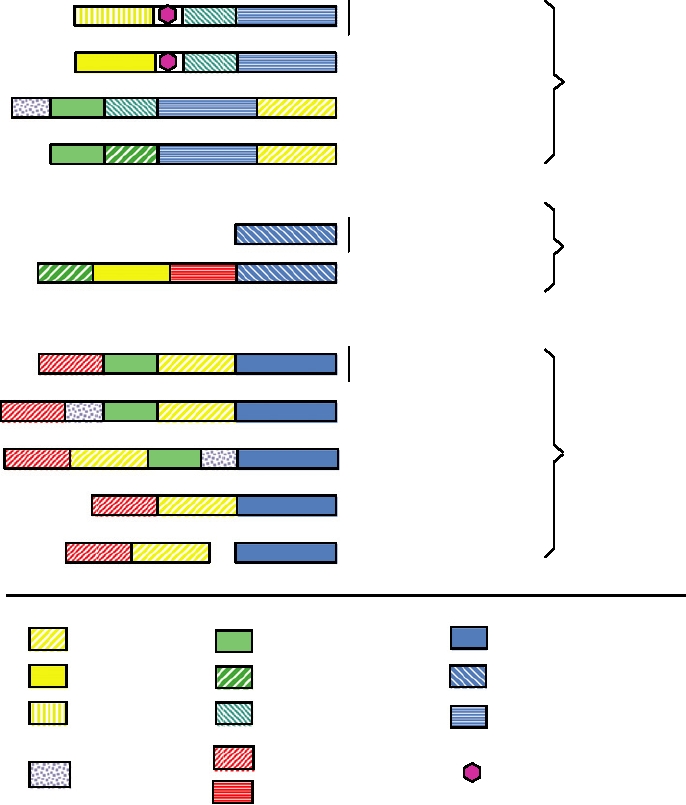
Picorna/Calici
Como/Nepoviruses
Potyviruses
Supergroup
1
Coronaviruses
Arteriviruses
Phage/Carmo/
Supergroup
Tombusviruses
2
Flaviviruses
Carla/Capillo/
Tymoviruses
Rubella
Supergroup
Alphaviruses
3
Tobamo/Tobravirus
Tricorna/Hordeivirus
Polymerase 3
Helicase 1
Papain protease
Polymerase 2
Helicase 2
Serine Protease
Helicase 3
Cysteine Protease
Polymerase 1
Methyltransferase 1
Vpg
X Protein
Methyltransferase 2
FIGURE 3.43 Genome organizations of plus-strand RNA viruses, grouped into three supergroups on the basis of
sequence relationships among the nonstructral proteins. The RNA polymerases (blue), the proteinases (green), and the
helicases (yellow) are each divided into three groups; the methyltransferases (red) are divided into two groups. From a
relatively small number of building blocks, it is possible to arrive at the genomes of all of these viruses by divergence of
individual domains and by recombination to reassemble them into different plans. Adapted from Strauss and Strauss (1994)
Figure 36.
picornavirus and in influenza virus (Chapter 4) were found
fill the same niche and selection ensures that the properties
to be 0.51% per year. Changes in third codon positions,
of the virus change only slowly. A second factor is that dif-
which are usually silent, occur more rapidly than changes
ferent domains of the genome, or even different nucleotides
in first or second codon positions, which usually result in
or amino acids, diverge at very different rates because of dif-
an amino acid substitution. In alphaviruses, which alternate
ferences in selection pressure. Studies of the rates of diver-
between insect and vertebrate hosts, the rate of divergence
gence of viruses perforce will measure the rates of domains
was significantly less, 0.03% per year, because changes that
that diverge most rapidly. There is no fossil record to tell
might be neutral or positively selected in one host are often
us when currently extant viruses might have diverged from
deleterious in the other host. One of the apparent paradoxes
one another, and viruses in collections have all been isolated
of such studies is the observation that despite rapid sequence
within the last 70 years. Thus, all studies of divergence in
divergence, the properties of most viruses appear to remain
nature examine only the divergence that has occurred within
stable for centuries or millennia. This is due in part to the fact
the last 70 years. Such considerations have two practi-
that although the sequence may drift, the virus continues to
cal implications. Vaccines developed against most viruses
Search WWH :