periods, and in stallions the virus may be secreted in semen
in which the genome is present in more than one segment,
for the life of the animal.
each segment is packaged separately into different particles
PRRSV causes respiratory distress in pigs of all ages
and infection requires the introduction into the same cell of at
and abortions and stillbirths in pregnant sows. SHFV is an
least one of each genome segment. It is of interest that such an
African virus that causes persistent, inapparent infections in
arrangement is common in plant viruses but nonexistent in ani-
African monkeys. When introduced into colonies of Asian
mal viruses, presumably because of differences in the mecha-
monkeys, however, it causes fatal hemorrhagic fever.
nisms by which plant and animal viruses spread and infect new
cells or new hosts. Many (+)RNA plant viruses are rod shaped,
formed using helical symmetry (e.g., tobacco mosaic virus, Fig.
FAMILY RONIVIRIDAE
2.2), while others are icosahedral (e.g., the comovirus cowpea
mosaic virus, Figs. 2.5 and 2.7). No (+)RNA plant viruses are
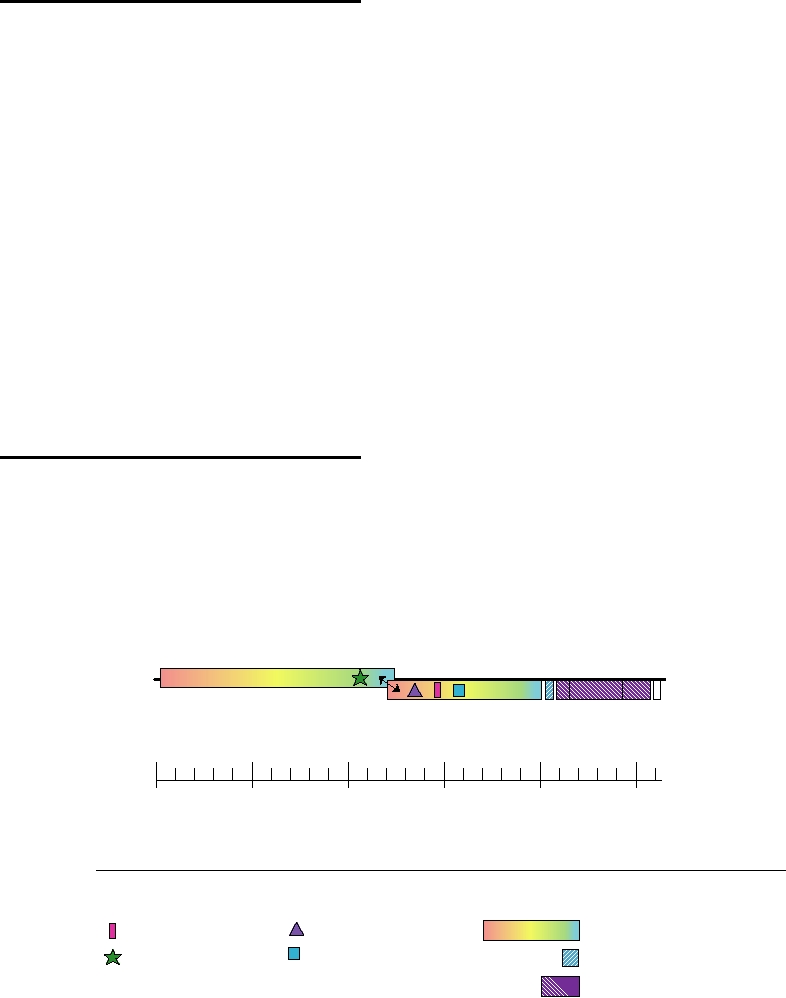
The Roniviridae, from rod-shaped nidovirus, are repre-
enveloped. Many of these viruses are major agricultural patho-
sented by a single known virus, gill-associated virus, which
gens responsible for a great deal of crop damage worldwide.
infects shrimp (Table 3.14). Its genome organization presents
Although important as plant pathogens, plant viruses will not
yet another permutation of how ancestral genes become associ-
be covered here except for a description of the genomes of cer-
ated with one another. The nonstructural genes, which occupy
tain families that are of particular interest because of what they
20 kb, are translated from the genomic RNA by mechanisms
tell us about the evolution of viruses.
that are very similar as those used by other members of the
Several families of (+)RNA plant viruses share sequence
Nidovirales (Fig. 3.40). However, the structural proteins are
identity with one another and with the alphaviruses. This
translated from only two subgenomic mRNAs, one that is trans-
collection of viruses, sometimes referred to as the Sindbis
lated into the nucleocapsid protein, and one that is translated
superfamily or the alphavirus superfamily, includes the
into a polyprotein precursor for the envelope proteins, which
alphaviruses, the tobamoviruses, the bromoviruses, and
are separated from one another by signalase. The assembled
other families of plant viruses. The genomes of the tobamo-
virion is bacilliform in shape, 150200 nm long and 4060 nm
virus tobacco mosaic virus (TMV), the bromovirus brome
in thickness. The virion thus resembles that of the rhabdovi-
mosaic virus (BMV), and the alphavirus Sindbis virus are
ruses (Chapter 4) rather than those of other nidoviruses.
compared in Fig. 3.41. The genome of TMV is one molecule
of (+)RNA and two subgenomic RNAs are produced. The
genome of BMV consists of three molecules of (+)RNA and
THE PLUS-STRAND RNA VIRUSES OF PLANTS
one subgenomic RNA is made. The alphaviruses have been
described. Notice that a characteristic of this superfamily
Most plant viruses possess (+)RNA as their genome. Some
is that all viruses in it produce at least one subgenomic
have as their genome a single RNA molecule and produce
mRNA. The members of this superfamily all share three
subgenomic mRNAs, whereas in others the viral genome is
proteins (or protein domains) with demonstrable sequence
divided into two or three or more segments. In plant viruses
homology, as indicated in the figure. These three are a viral
ORF1A
5'
ORF4 3' ORFs
ORF1B
ORF2 ORF3
(A)n
-1 Frameshift site
p20 gp116 gp64
PROTEINS
0
5
10
15
20
25
kb
Coding Domains
Enzyme Motifs
Polymerase (GDD)
Nonstructural proteins
Zinc finger
Helicase
3C-like protease
Nucleocapsid protein
Virion transmembrane
proteins
FIGURE 3.40 Genome organization of the Roniviridae. Redrawn from Cowley and Walker (2002).
RNAs
(All
Strand)
Tobamovirus
CAP
Genome RNA
CAP
mRNAs
C
CAP
Bromovirus
RNA 2
RNA 1
RNA 3
Genome RNAs
CAP
CAP
CAP
mRNA
C
CAP
Alphavirus
An Genome RNA
CAP
nsP2
nsP1
nsP3
nsP4
An
CAP
STRUCTURAL PROTEINS
mRNA
Methyltransferase
Polymerase
Plant virus "movement" protein
Helicase
Papain protease
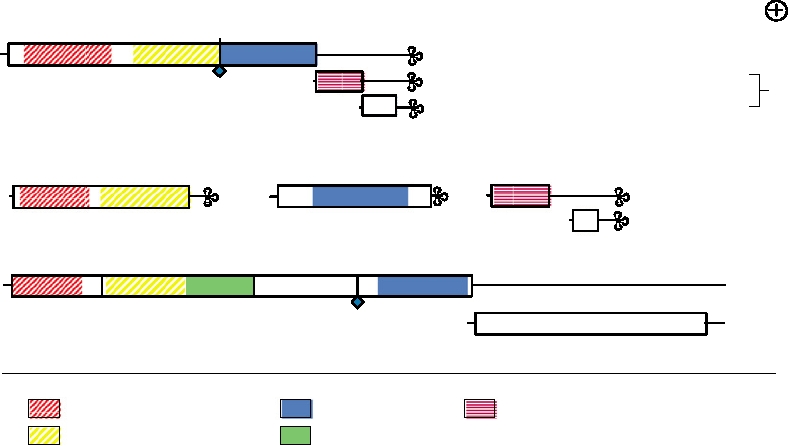
FIGURE 3.41 Comparison of the genome organization of alphavirus Sindbis with representatives of two plant virus
families. Three shaded domains illustrate regions of low but significant sequence homology, which extend over hundreds
of amino acids, within the methyltransferase, helicase, and polymerase proteins. The blue diamond is a leaky termination
codon that is read through to produce the downstream blue-shaded domains in the tobamoviruses and the alphaviruses.
C is the coat protein. The plant viruses have no module corresponding to the protease in nsP2 nor to protein nsP3. The
alphaviruses have no domain corresponding to the "movement" protein of plant viruses. Adapted from Strauss and Strauss
(1994), Figure 35.
RNA polymerase, a helicase, and a capping enzyme (char-
but is not enveloped. Thus, recombination has brought
acterized by methyltransferase activity). In the case of the
together different RNA replication modules with different
alphaviruses and the tobamoviruses, all three domains are
structural protein modules to give rise to the current fami-
found on one genome segment and readthrough is required
lies of viruses.
to translate the polymerase. In the bromoviruses, the cap-
Similar considerations pertain to two families of plant
ping enzyme and the helicase are encoded on one segment,
viruses (the Comoviridae and the Potyviridae) and the ani-
but the polymerase is encoded on a different segment. The
mal picornaviruses, which are all related to one another and
alphaviruses encode a protease to separate the three domains
are sometimes referred to as the picornavirus superfamily.
from one another, but the plant viruses do not. While these
The Comoviridae have a bipartite genome, whereas the
three shared proteins have clearly diverged from a common
Potyviridae and the Picornaviridae have a single molecule
ancestral source, other domains within the nonstructural
of RNA as their genome. Characteristics of this superfamily
include the absence of subgenomic RNAs, the presence of 5′
proteins are different from family to family. The alphavi-
VPg and 3′ poly(A) on the viral RNAs, and the production
rus protease and nsP3 are not shared with the plant viruses,
while the plant viruses possess movement proteins that are
of at least one protease. The genome organizations of two
not shared with the alphaviruses. The structural proteins
members of the Comoviridae that belong to different gen-
of the different families are also distinct. These observa-
era, tomato black ring virus (genus Nepovirus) and cowpea
tions clearly point to the occurrence of extensive recom-
mosaic virus (genus Comovirus), are compared with that of
binational events during the evolution of this group of
picornavirus poliovirus in Fig. 3.42. The members of this
viruses from a common ancestral source. Recombination
superfamily have demonstrable homologies in their RNA
polymerases, 2C helicases, and 3Cpro proteases. Further,
has brought together new combinations of genes appropri-
the RNA genomes have a 5′ VPg and are polyadenylated,
ate to the different lifestyles of the various members of
the superfamily. In addition, the structural proteins differ
as noted. Proteases, VPg's, and poly(A) are very unusual in
among these three families so that the structures of the vir-
plant viruses, found only in members of this superfamily. It
ions are very different from one another. The alphaviruses
is clear that these viruses are all related to one another, and
are enveloped, icosahedral particles (Figs. 2.5, 2.14A, and
that multiple recombination events have taken place to give
2.25C). TMV is rod shaped (Fig. 2.2). BMV is icosahedral
rise to the current families.
Search WWH :