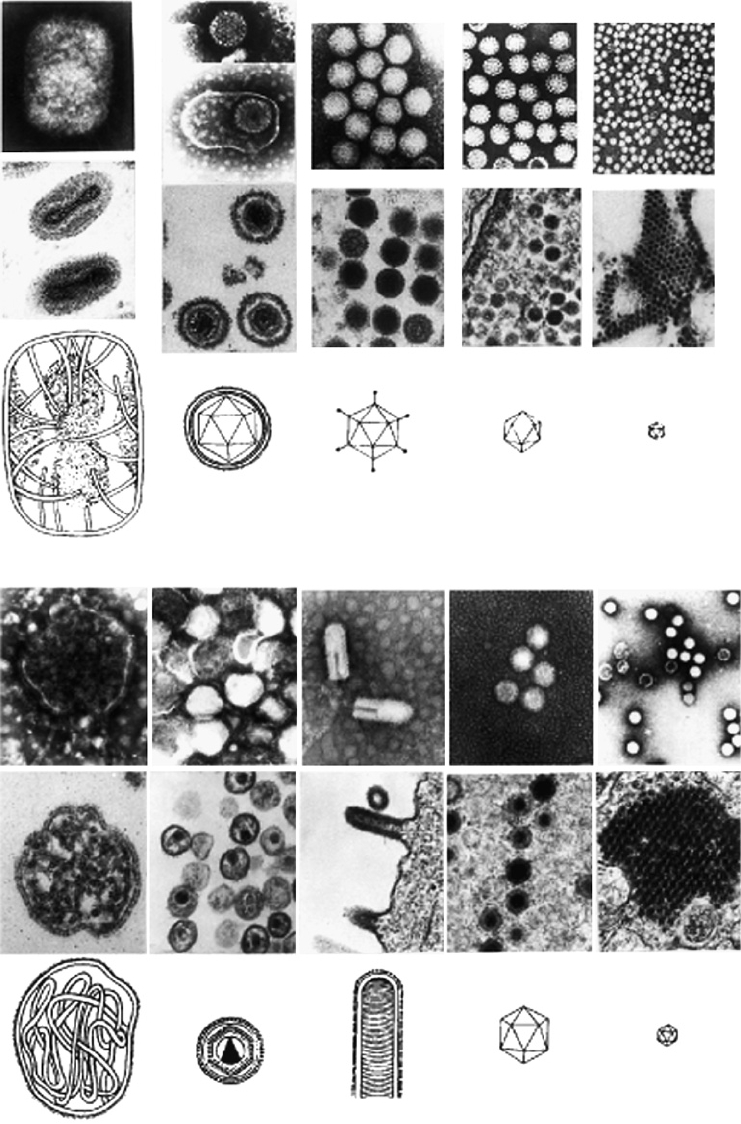
TABLE 2.1
Listing of Selected Vertebrate Virus Families by Morphology
Morphology
Triangulation
Morphology
of particle/virus family
Enveloped
number
of nucleocapsid
Figure numbers
Icosahedral
Adenoviridae
No
T = 25
Not applicable
Figs. 2.1, 2.12
T = 13l a
Reoviridae
No
Icosahedral
Figs. 2.1, 2.5, 2.11
P = 7d a,b
Figs. 2.1,c 2.5
Papillomaviridae
No
Not applicable
P = 7d a,b
Figs. 2.1,c 2.5, 2.10
Polyomaviridae
No
Not applicable
Parvoviridor
No
T=1
Not applicable
Figs. 2.1, 2.5
Figs. 2.1,d 2.5, 2.7, 2.8
P = 3b
Picornaviridae
No
Not applicable
Astroviridae
No
Not applicable
Caliciviridae
No
T=3
Not applicable
Herpesviridae
Yes
T = 16
Icosahedral
Figs. 2.1, 2.5, 2.20
Togaviridae
Yes
T=4
Icosahedral
Figs. 2.5, 2.14, 2.18
Flaviviridae
Yes
T=3
Icosahedral
Figs. 2.5, 2.18
Irregular
Poxviridae (ovoid)
Yes
Dumbbell
Figs. 2.1, 2.24
Rhabdoviridae (bacilliform)
Yes
Coiled helix
Figs. 2.1, 2.23
Spherical
Retroviridae
Yes
?
Icosahedral?
Figs. 2.1, 2.21
Rounde
Coronaviridae
Yes
Helical or tubular
Paramyxoviridae
Yes
Helical
Fig. 2.22
Orthomyxoviridae
Yes
Helical
Figs. 2.1, 2.22
Bunyaviridae
Yes
Helical
Arenaviridae
Yes
Helical
Hepadnaviridae
Yes
T=3&4
Icosahedral
Filamentousc
Filoviridae
Yes
Helical
Fig. 2.19
a
Two mirror image structures can be formed using T = 13 or T = 7, a symmetry referred to as "d" or "l."
b
Because the subunits are not exactly equivalent, Papillomaviridae, Polyomaviridae, as well as poliovirus, have "pseudo-triangulation numbers" so are
referred to as P = 7, P = 7, and P = 3 symmetries respectively.
c
Papovaviridae, referred to in Fig. 2.1 have now been separated into Polyomaviridae and Papillomaviridae.
d
Enterovirus, referred to in Fig. 2.1 is a genus of the Picornaviridae.
e
Virions are often pleiomorphic.
ICOSAHEDRAL SYMMETRY
edge possesses twofold rotational symmetry. Thus the
icosahedron is characterized by twofold, threefold, and five-
fold symmetry axes. The dodecahedron, the next simpler
Virions can be approximately spherical in shape, based
regular solid, has the same symmetry axes as the icosahe-
on icosahedral symmetry. Since the time of Euclid, there
dron and is therefore isomorphous with it in symmetry: the
have been known to exist only five regular solids in which
dodecahedron has 12 faces which are regular pentagons, 20
each face of the solid is a regular polygon: the tetrahedron,
vertices where three faces meet, and 30 edges with twofold
the cube, the octahedron, the dodecahedron, and the icosa-
symmetry. The three remaining regular solids have differ-
hedron. The icosahedron has 20 faces, each of which is a
ent symmetry axes. The vast majority of regular viruses that
regular triangle, and thus each face has threefold rotational
appear spherical have icosahedral symmetry.
symmetry (Fig. 2.3A). There are 12 vertices where 5 faces
In an icosahedron, the smallest number of subunits that
meet, and thus each vertex has fivefold rotational symme-
can form the three-dimensional structure is 60 (5 subunits at
try. There are 30 edges in which 2 faces meet, and each
A
POX-
HERPES-
ADENO-
PAPOVA-
PARVO-
B
MYXO-
RETRO-
RHABDO-
REO-
ENTERO-
FIGURE 2.1 Relative sizes and shapes of representative (A) DNA- and (B) RNA- containing viruses. In each panel the
top row shows negatively stained virus preparations, the second row shows thin sections of virus-infected cells, and the
bottom row illustrates schematic diagrams of the viruses. Magnification of the electron micrographs is 50,000. From
Granoff and Webster (1999), p. 401.
RNA
100 nm
A
B
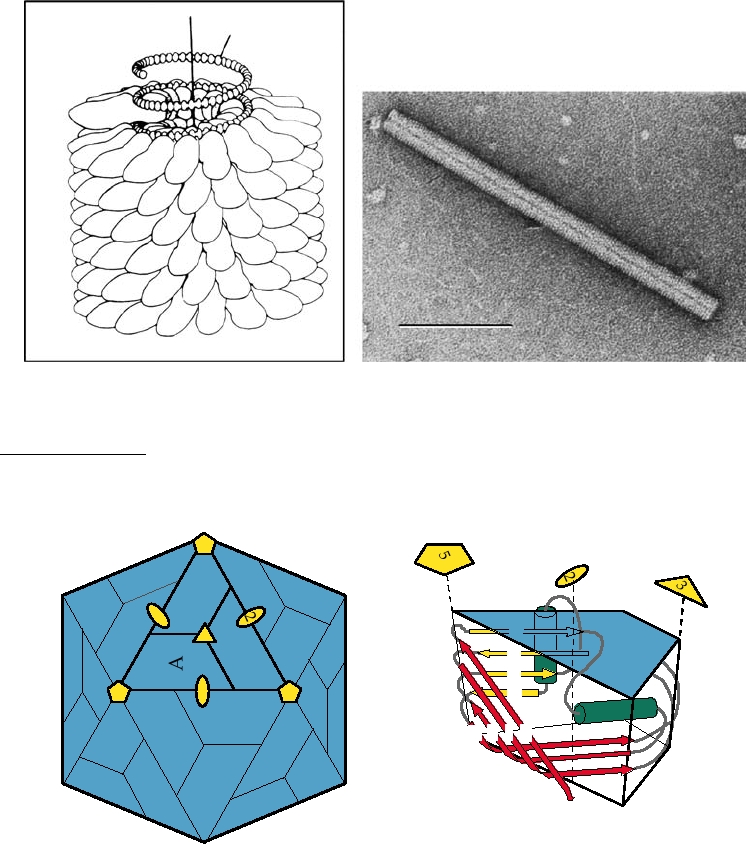
FIGURE 2.2 Structure of TMV, a helical plant virus. (A) Schematic diagram of a TMV particle showing about 5%
of the total length. From Murphy et al. (1995), p. 434. (B) Electron micrograph of a negatively stained TMV rod. From
www.ncbi.nlm.nih.gov.
5
A
2
C
3
A
H
E
HOOC
2
F
5
5
GD I B
NH
A
B
FIGURE 2.3 A simple icosahedral virus. (A) Diagram of an icosahedral capsid made up of 60 identical copies of
a protein subunit, shown as blue trapezoids labeled "A." The twofold, threefold, and fivefold axes of symmetry are
shown in yellow. This is the largest assembly in which every subunit is in an identical environment. (B) Schematic
representation of the subunit building block found in many RNA viruses, known as the eightfold β barrel or β sandwich.
The β sheets, labeled B through I from the N terminus of the protein, are shown as yellow and red arrows; two possible
α helices joining these sheets are shown in green. Some proteins have insertions in the CD, EF, and GH loops, but
insertions are uncommon at the narrow end of the wedge (at the fivefold axis). From Granoff and Webster (1999) Vol. 3,
color plate 31. [Originally from J. Johnson (1996)].
each of the 12 vertices, or viewed slightly differently, 3 units
1, 3, 4, 7, 9, 12, 13, 16, and so forth. A subunit defined in
on each of the 20 triangular faces). Some viruses do in fact
this way is not necessarily formed by one protein molecule,
use 60 subunits, but most use more subunits in order to pro-
although in most cases this is how a structural subunit is in
vide a larger shell capable of holding more nucleic acid.
fact formed. Some viruses that form regular structures that
The number of subunits in an icosahedral structure is 60T,
are constructed using icosahedral symmetry principles do
where the permissible values of T are given by T = H 2 +
not possess true icosahedral symmetry. In such cases they
HK + K 2, where H and K are integers and T is called the
are said to have pseudo-triangulation numbers. Examples
triangulation number. Permissible triangulation numbers are
are described later.
Structural studies of viruses have shown that the cap-
Because the size of the icosahedral shell is fixed by
sid proteins that form the virions of many plant and animal
geometric constraints, it is difficult for a change in the size
icosahedral viruses have a common fold. This fold, an eight-
of a viral genome to occur. A change in size will require a
stranded antiparallel β sandwich, is illustrated in Fig. 2.3B.
change in the triangulation number or changes in the cap-
The presence of a common fold suggests that these capsid
sid proteins sufficient to produce a larger or smaller internal
proteins have a common origin even if no sequence identity is
volume. In either case, the changes in the capsid proteins
detectable. The divergence in sequence while maintaining this
required are relatively slow to occur on an evolutionary
basic fold is illustrated in Fig. 2.4, where capsid proteins of
timescale and the size of an icosahedral virus is "frozen" for
three viruses are shown. SV40 (family Polyomaviridae), polio-
long periods of evolutionary time. For this reason, as well as
virus (family Picornaviridae), and bluetongue virus (family
for other reasons, most viruses have optimized the informa-
Reoviridae) are a DNA virus, a single-strand RNA virus, and
tion content in their genomes, as will be clear when indi-
a double-strand RNA virus, respectively. Their capsid pro-
vidual viruses are discussed in the following chapters.
teins have insertions into the basic eight-stranded antiparallel
β-sandwich structure that serve important functions in virus
Comparison of Icosahedral Viruses
assembly. However, they all possess a region exhibiting the
common β-sandwich fold and may have originated from a
Cryoelectron microscopy has been used to determine the
common ancestral protein. Thus, once a suitable capsid protein
structure of numerous icosahedral viruses to a resolution of
arose that could be used to construct simple icosahedral parti-
7 to 25 Å. For this, a virus-containing solution on an electron
cles, it may ultimately have been acquired by many viruses.
microscope grid is frozen very rapidly so that the sample
The viruses that possess capsid proteins with this fold may be
is embedded in amorphous frozen water. The sample must
related by descent from common ancestral viruses, or recombi-
be maintained at liquid nitrogen temperatures so that ice
nation may have resulted in the incorporation of this successful
crystals do not form and interfere with imaging. Unstained,
ancestral capsid protein into many lines of viruses.
slightly out-of-focus images of the virus are captured on
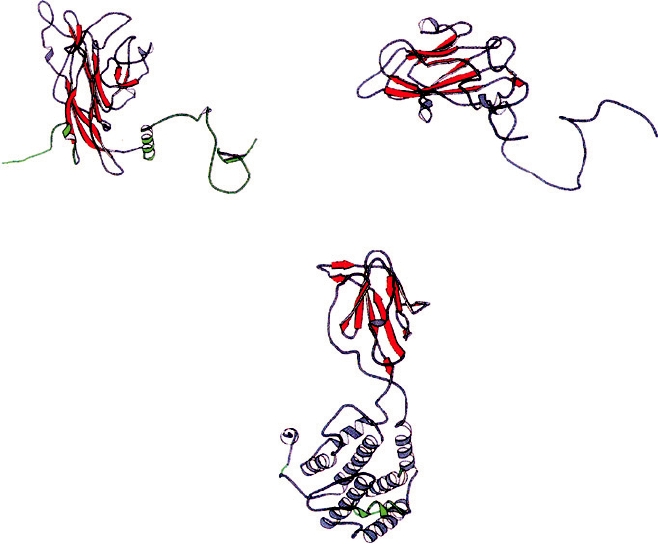
C(361)
C(238)
N(1)
N(15)
SV40 VP1 P = 7
Poliovirus1 VP3 P = 3
C(349)
N(1)
Bluetongue virus VP7 T = 13
FIGURE 2.4
Structure of three vertebrate virus protein subunits that assemble into icosahedral shells. The N termini
and C termini are labeled with the residue number in parenthesis. The β barrels are shown as red arrows, α helices are
gray coils, and the subunit regions involved in quasi-symmetric interactions that are critical for assembly are colored
green. SV40 and PV have triangulation numbers of "pseudo-T=7" or P = 7 and "pseudo-T=3" or P = 3, respectively.
Adapted from Granoff and Webster (1999), Vol. 3, plate 32.
film, or more recently captured electronically, using a low
which is 1250 Å in diameter and has T=16 symmetry (the
dose of electrons. These images are digitized and the den-
virion is enveloped but only the nucleocapsid is regular).
sity measured. Mathematical algorithms that take advantage
The rotavirus and reovirus virions are smaller and have
of the symmetry of the particle are used to reconstruct the
T=13. Human papillomavirus and mouse polyomavirus are
structure of the particle.
pseudo-T=7. Ross River virus (RRV) (family Togaviridae)
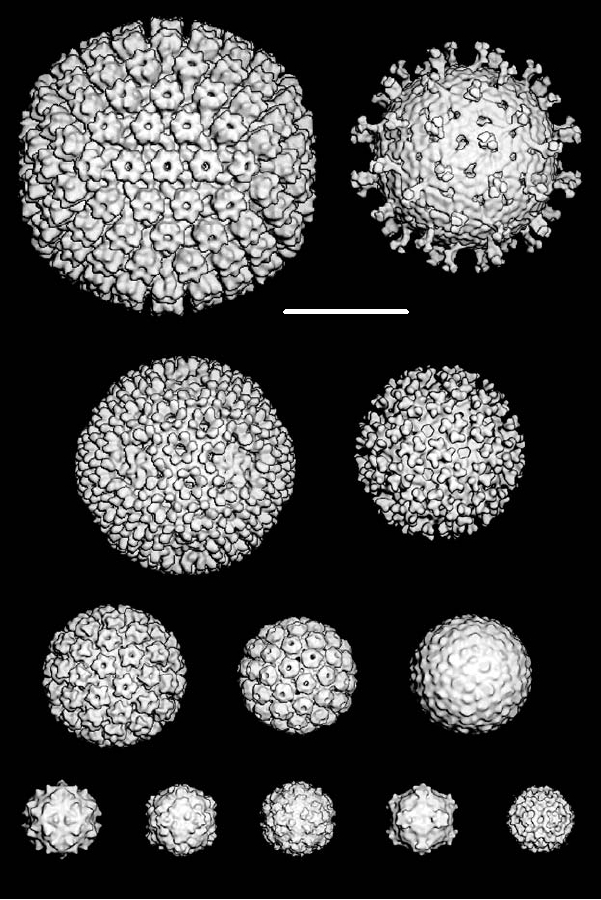
A gallery of structures of viruses determined by cryoelec-
is enveloped but has regular symmetry, with T=4. Several
tron microscopy is shown in Fig. 2.5. All of the images are to
examples of viruses with T=3 or pseudo-T=3 are shown
scale so that the relative sizes of the virions are apparent. The
(dengue 2, flock house, rhino-, polio-, and cowpea mosaic
largest particle is the nucleocapsid of herpes simplex virus,
viruses, of which dengue 2 is enveloped but regular and the
Rotavirus
(1000 Å)
Herpes Simplex
500 Å
(1250 Å)
Ross River (700 Å)
Reovirus (Lang)
(850 Å)
Dengue 2 (490 Å)
Human papilloma (600 Å)
Polyoma (495 Å)
Flockhouse Human rhinovirus
Poliovirus Cowpea mosaic
B19 parvovirus
(330 Å)
(320 Å)
(320 Å)
(312 Å)
(260 Å)
FIGURE 2.5 Gallery of three-dimensional reconstructions of icosahedral viruses from cryoelectron micrographs. All
virus structures are surface shaded and are viewed along a twofold axis of symmetry except Ross River, which is viewed
along a three-fold axis. All of the images are of intact virus particles except for the herpes simplex structure, which is of
the nucleocapsid of the virus. Most of the images are taken from Baker et al. (1999), except the images of Ross River
virus and of dengue virus, which were kindly provided by Drs. R. J. Kuhn and T. S. Baker.
construct the shell. The structures of two insect viruses
rest are not enveloped). B19 parvovirus has T=1. The gen-
that are also simple T=3 structures have also been solved.
eral correlation is that larger particles are constructed using
As an example of these simple structures, the T=3 capsid
higher triangulation numbers, which allows the use of larger
of the insect virus, flock house virus (family Nodaviridae),
numbers of protein subunits. Larger particles accommodate
is illustrated in Fig. 2.6.
larger genomes.
The 180 subunits in these T=3 structures interact with one
another in one of two different ways, such that the protein
Atomic Structure of T=3 Viruses
shell can be thought of as being composed of an assembly
of 60 AB dimers and 30 CC dimers (Fig. 2.6A). The bond
Because the simplest viruses are regular structures, they
angle between the two subunits of the dimer is more acute
will often crystallize, and such crystals may be suitable for
in the AB dimers than in the CC dimers (Figs. 2.6B and C).
X-ray diffraction. Many viruses formed using icosahedral
For the plant viruses, there are N-terminal and C-terminal
symmetry principles have been solved to atomic resolu-
extensions from the capsid proteins that are involved in
tion, and a discussion of representative viruses that illus-
interactions between the subunits and with the RNA. The
trate the principles used in construction of various viruses
N-terminal extensions have a positively charged, disordered
is presented here.
domain for interacting with and neutralizing the charge on
Among T=3 viruses, the structures of several plant
the RNA and a connecting arm that interacts with other
viruses, including tomato bushy stunt virus (TBSV) (genus
subunits. In the case of the CC dimers, the connecting arms
Tombusvirus, family Tombusviridae), turnip crinkle virus
interdigitate with two others around the icosahedral three-
(TCV) (genus Carmovirus, family Tombusviridae), and
fold axis to form an interconnected internal framework. In
Southern bean mosaic virus (SBMV) (genus Sobemovirus,
the case of the AB conformational dimer, the arms are dis-
not yet assigned to family), have been solved. All three
ordered, allowing sharper curvature. For flock house virus,
of these viruses have capsid proteins possessing the
eight-stranded antiparallel β sandwich. T=3 means that
the RNA plays a role in controlling the curvature of the CC
dimers, as illustrated in Fig. 2.6.
180 identical molecules of capsid protein are utilized to
A.
B.
C5
A
5
B
B
dsRNA
Arm
B2
dsRNA
2
A
C.
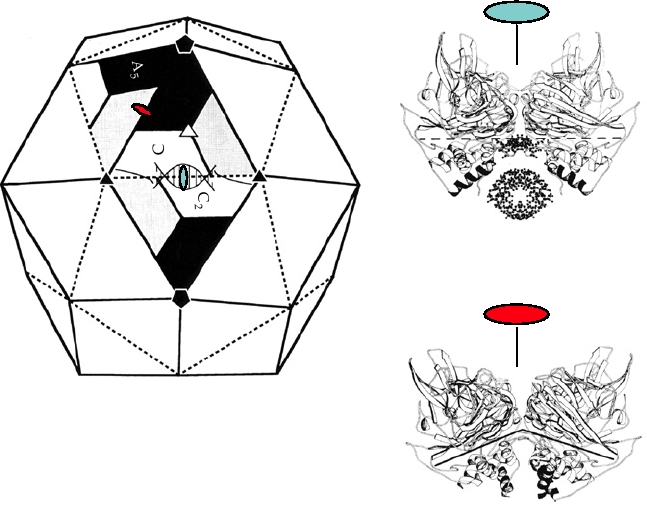
FIGURE 2.6 (A) Diagrammatic representation of a T=3 virus, flock house virus. The positions of the three identical
proteins that make up a triangular face are only quasi-equivalent. The angle between the A and B5 units (shown with a
red oval and in diagram (C) is more acute than that along the CC2 edge, shown with a blue oval, and diagram (B). This
difference in the angles is due to the presence of an RNA molecule located under the CC2 edge. From Johnson (1996),
with permission.
Atomic Structure of Viruses Having
Pseudo-T=3 Symmetry
The structures of several picornaviruses and of a plant
comovirus (cowpea mosaic virus) have also been solved to
atomic resolution. The structures of these viruses are simi-
lar to those of the plant T=3 viruses, but the 180 subunits
that form the virion are not all identical. A comparison of
the structure of a T=3 virus with those of poliovirus and of
cowpea mosaic virus is shown in Fig. 2.7. Poliovirus has
60 copies of each of three different proteins, whereas the
comovirus has 60 copies of an L protein (each of which
fills the niche of two units) and 60 copies of an S protein.
All three poliovirus capsid proteins have the eight-stranded
antiparallel β-sandwich fold. In the comoviruses, the L pro-
tein has two β-sandwich structures fused to form one large
protein, and the S protein is formed from one sandwich.
The structures of the picornavirus and comovirus virions
are called pseudo-T=3 or P=3, since they are not true T=3
structures.
The picornavirus virion is 300 Å in diameter. The 60 mol-
ecules of each of the three different proteins have different
roles in the final structure, as illustrated in Fig. 2.8, in which
FIGURE 2.8
Three-dimensional space-filling model of the human
the structure of a rhinovirus is shown. Notice that five copies
rhinovirus 14 virion, based upon X-ray crystallographic data. VP1 is shown
in blue, VP2 in green, and VP3 in red. VP4 is interior and not visible in this
of VP1 are found at each fivefold axis (compare Fig. 2.7 with
view. This figure was kindly provided by Dr. Michael Rossmann.
Fig. 2.8). VP1, VP2, and VP3 are structurally related to one
another, as stated, all possessing the common β-sandwich
fold. There exists a depression around each fivefold axis of
rhinoviruses that has been termed a "canyon." This depres-
Atomic Structure of Polyomaviruses
sion is believed to be the site at which the virus interacts with
The structures of both mouse polyomavirus and of
the cellular receptor during entry, as illustrated in Fig. 2.9.
SV40 virus, two members of the family Polyomaviridae,
This interaction is thought to lead to conformational changes
have been solved to atomic resolution. Both viruses pos-
that open a channel at the fivefold axis, through which VP4
sess pseudo-T=7 icosahedral symmetry. Although T=7
is extruded, followed by the viral RNA.
T=3
Poliovirus
Cowpea Mosaic Virus
S
A
S
VP1
A
L
VP1
VP3
L
B
VP2
A
C
S
VP1
VP2
B
L
C
L
B
C
VP3
VP2
VP3
B
C
L
L
A
VP1
VP3
C
VP2
VP2
B
S
VP3
A
S
VP1
L
L
C
L
B
A
VP3
VP2
L
S
VP1
S
B
C
VP2
VP3
VP1
A
B
C
VP2
VP3
C
A
B
VP1
S
VP2
L
VP3
L
A
C
VP1
S
VP2
A
B
S
VP1
VP3
L
L
CA
A
S
B
VP2 VP1 VP1 VP3
S
B
C
VP3 VP2
L
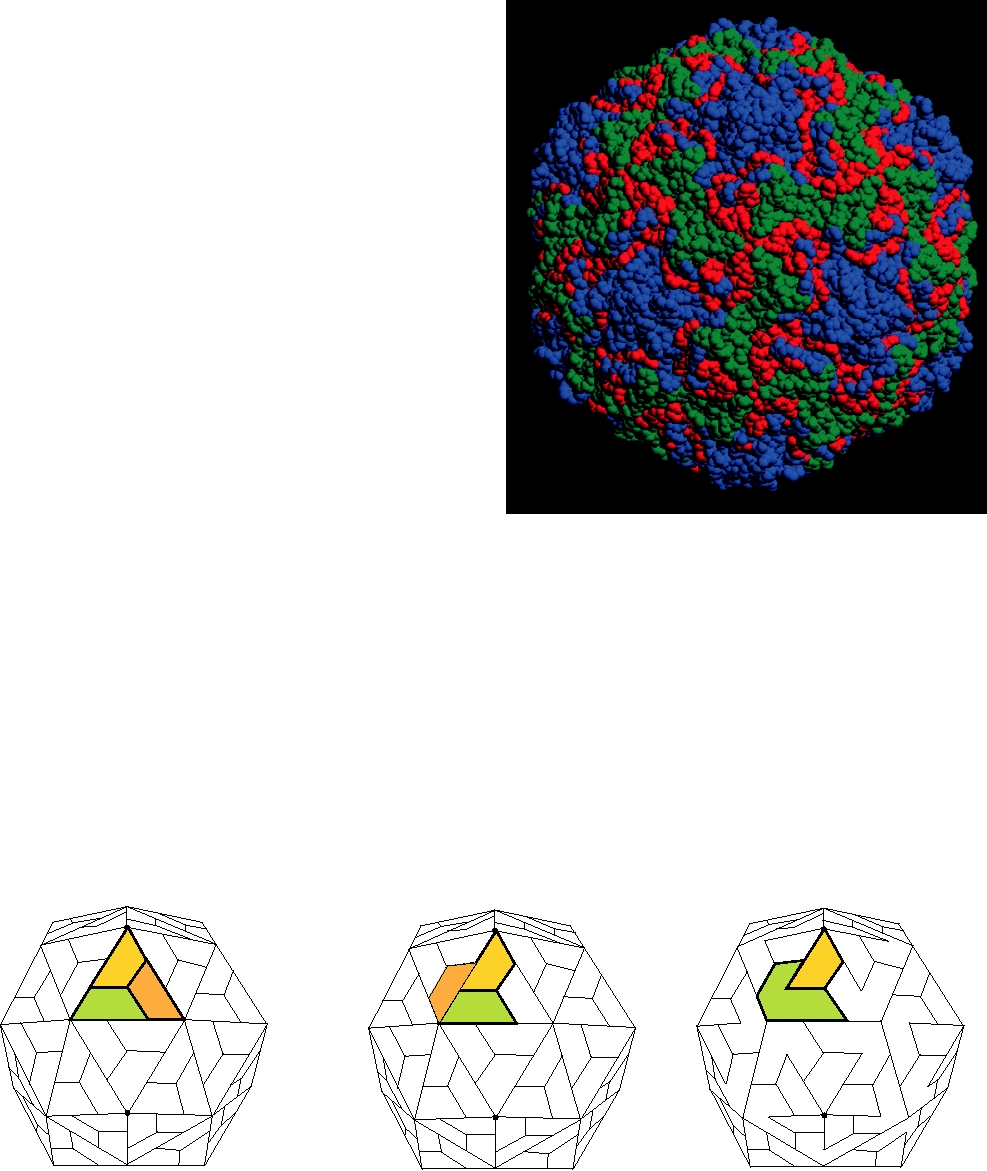
FIGURE 2.7 Arrangement of the coat protein subunits of comoviruses compared with those of simple T=3 viruses and
picornaviruses. In simple viruses, the asymmetric unit contains three copies of a single protein β sandwich, labeled A, B,
and C in order to distinguish them. In picornaviruses such as poliovirus the asymmetric unit is made up of three similar
but not identical proteins, all of which have the β-sandwich structure. In comoviruses such as cowpea mosaic virus, two
of the β-sandwich subunits are fused to give the L protein. Adapted from Granoff and Webster (1999), p. 287.
ICAM-1
D2
VP1
"Canyon''
D1
VP2
Drug-binding pocket
VP3 bcylinder
VP3
VP4
RNA
Five-fold Axis
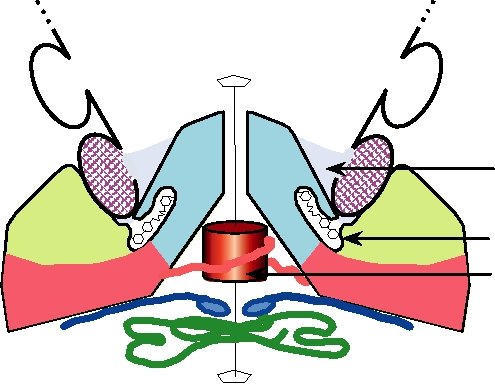
FIGURE 2.9 Binding of the rhinovirus receptor, ICAM-1, to the "canyon" at a fivefold vertex of a virion of a major
group human rhinovirus. The colors of the three virion proteins are the same as those shown in the surface view in Fig.
2.8. The distal two domains of ICAM-1 are represented schematically (cross-hatched) as they were in Fig. 1.5. The
amino-terminal domains of the five VP3 molecules around the fivefold axis form a five-stranded β cylinder on the virion's
interior and are thought to stabilize the pentamer. Below the canyon is the hydrophobic pocket where certain antiviral
drugs (indicated schematically in black) are known to bind. Adapted from Kolatkar et al. (1999).
symmetry would require 420 subunits, these viruses con-
solve the structure of one or more members of three gen-
tain only 360 copies of a major structural protein known
era within the Reoviridae, namely Reovirus, Rotavirus, and
as VP1. These 360 copies are assembled as 72 pentamers.
Orbivirus, to about 25-Å resolution. Structures of a reovirus
Twelve of the 72 pentamers lie on the fivefold axes and
and of a rotavirus are shown in Fig. 2.5. The complete struc-
the remaining 60 fill the intervening surface in a closely
ture of virions has not been determined because of their large
packed array (Fig. 2.10). These latter pentamers are thus
size, but in a remarkable feat the atomic structure of the core
sixfold coordinated and the proteins in the shell are not all
of bluetongue virus (genus Orbivirus) has now been solved.
in quasi-equivalent positions, a surprising finding for our
This is the largest structure determined to atomic resolu-
understanding of the principles by which viruses can be
tion to date. Solution of the structure was possible because
constructed. The pentamers are stabilized by interactions
the virus particle had been solved to 25 Å by cryoelectron
of the β sheets between adjacent monomers in a pentamer
microscopy, and the structures of a number of virion proteins
(Fig. 2.10C). The pentamers are then tied together by
had been solved to atomic resolution by X-ray diffraction.
C-terminal arms of VP1 that invade monomers in an adja-
Fitting the atomic structure of the proteins into the 25-Å
cent pentamer (Figs. 2.10B and C). Because each pentamer
structure gave a preliminary reconstruction at high resolu-
that is sixfold coordinated has five C-terminal arms to
tion, which allowed the interpretation of the X-ray data to
interact with six neighboring pentamers, the interactions
atomic resolution.
between monomers in different pentamers are not all iden-
Core particles are formed following infection, when the
tical (Fig. 2.10B). Flexibility in the C-terminal arm allows
outer layer is proteolytically cleaved (described in more
it to form contacts in different ways.
detail in Chapter 5). The structure of the inner surface of
the bluetongue virus core is shown in Fig. 2.11A and of the
outer surface in Fig. 2.11B. The outer surface is formed by
Atomic Structure of Bluetongue Virus
780 copies of a single protein, called VP7, in a regular T=13
Members of the reovirus family are regular T=13 icosahe-
icosahedral lattice. The inner surface is surprising, however.
dral particles. They are composed of two or three concentric
It is formed by 120 copies of a single protein, called VP3.
protein shells. Cryoelectron microscopy has been used to
These 120 copies have been described as forming a T=2
A
B
A
B
b
C
F
5
a,
g
a
5
3
2
a,,
g
5
E
D
C
Pentamer axis
b sheet from adjacent
VP1 in same pentamer
Red and blue arrows are b sheets
C-terminal domain from an
adjacent pentamer
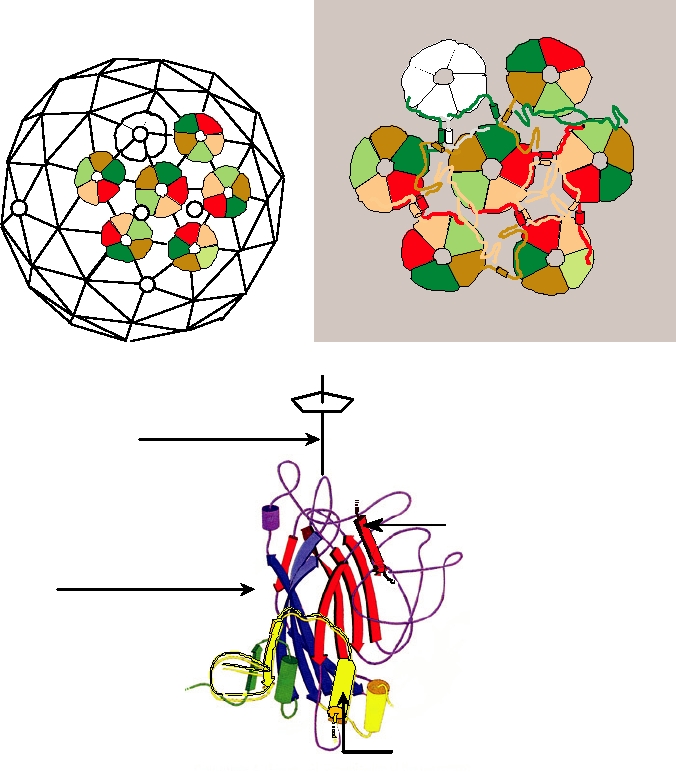
FIGURE 2.10 Organization of the capsid of the polyomavirus SV40. (A) Arrangement of the strict pentamers (white)
and quasi pentamers (colored) on the T=7d icosahedral lattice. (B) Schematic showing the pattern of interchange of arms
in the virion. The central pentamer shares "arms" with six neighboring pentamers. (C) A single VP1 subunit, viewed
normal to the pentamer axis. The N-terminal domain is green, the C-terminal domain is yellow, and the β sandwich is
shown as arrows of blue and red. The central yellow C-terminal domain (outlined in black) comes from an adjacent
pentamer and the β sheet outlined in black comes from the neighboring VP1 within the same pentamer. From Figure 1
Stehle et al. (1996) and Fields et al. (1996), Color Plate 4.
Structure of Adenoviruses
lattice. Because T=2 is not a permitted triangulation number,
these 120 copies, strictly speaking, form a T=1 lattice in
Cryoelectron microscopy has also been applied to adeno-
which each unit of the lattice is composed of two copies of
viruses, which have a triangulation number of 25 or pseudo-
VP3. However, the interactions are not symmetrical, leading
25. Various interpretations of the structure of adenoviruses,
to the suggested terminology of T=2.
both schematic and as determined by microscopy or crystal-
It has been suggested that the inner core furnishes a template
lography, are shown in Fig. 2.12. Three copies of a protein
for the assembly of the T=13 outer surface. The reasoning is
called the hexon protein associate to form a structure called a
that a T=13 structure may have difficulty in forming, whereas
hexon (Fig. 2.12C). The hexon is the basic building block of
the T=2 (or T=1) structure could form readily. In this model,
adenoviruses. Five hexons, called peripentonal hexons, sur-
the threefold symmetry axis of the inner surface could serve to
round each of the 12 vertices of the icosahedron (which, as has
nucleate VP7 trimers and organize the T=13 structure.
been stated, have fivefold rotational symmetry). Between the
Search WWH :