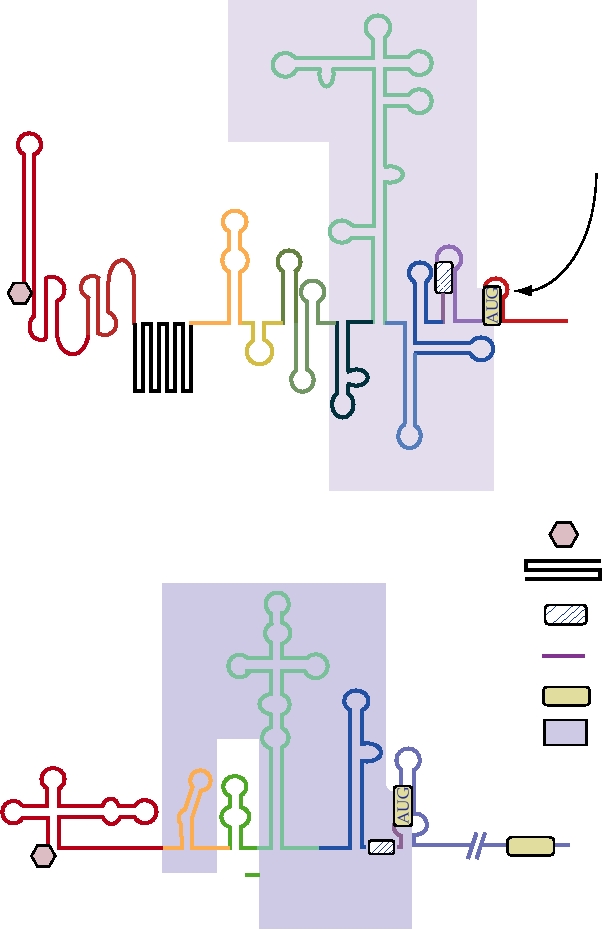
which is circular and largely double stranded. Thus their
of the genome. Genera are grouped into families, which
genome replicates through an RNA intermediate (DNA →
can be considered the fundamental unit of virus taxonomy.
RNA → DNA). Just as the minus-strand RNA viruses and
Classification into families is based on the type and size of
double-strand RNA viruses package their replicase proteins,
the nucleic acid genome, the structure of the virion, and the
the retroviruses package active RT, which is required
strategy of replication used by the virus, which is determined
to begin the replication of the genome in the virions.
in part by the organization of the genome. Groupings into
Although in many treatments the retroviruses are
families are not always straightforward because little or no
considered with the RNA viruses and the hepadnaviruses
sequence identity is present between members of different
with the DNA viruses, we consider these viruses to form
genera. However, uniting viruses into families attempts to
a distinct class, the RT-encoding class, and in this topic
recognize evolutionary relationships and is valuable for
references to RNA viruses or to DNA viruses are not meant
organizing information about viruses.
to apply to the retroviruses or the hepadnaviruses.
Higher taxonomic classifications have not been recognized
All viruses, with one exception, are haploid; that is, they
for the most part. To date only three orders (Caudovirales,
contain only one copy of the genomic nucleic acid. The
Nidovirales, Mononegavirales) have been established that
exception is the retroviruses, which are diploid and contain
group together a few families. Taxonomic classification at
two identical copies of the single-stranded genomic RNA.
higher levels is difficult because viruses evolve rapidly and
The nucleic acid genome may consist of a single piece of
it can be difficult to prove that any two given families are
DNA or RNA or may consist of two or more nonidentical
descended from a common ancestor, although it is almost
fragments. The latter can be considered analogous to chro-
certain that higher groupings based on common evolution
mosomes and can reassort during replication. In the case
do exist and will be elucidated with time. Viral evolution
of animal viruses, when a virus has more than one genome
involves not only sequence divergence, however, but also the
segment, all of the different segments are present within a
widespread occurrence of recombination during the rise of the
single virus particle. In the case of plant viruses with mul-
modern families, a feature that blurs the genetic relationships
tiple genome segments, it is quite common for the different
between viruses. Two families may share, for example, a
genome segments to be separately encapsidated into differ-
related polymerase gene but have structural protein genes that
ent particles. In this case, the infectious unit is multipartite:
appear unrelated; how should such viruses be classified?
Infection to produce a complete replication cycle requires
The ICTV has recognized 5450 viruses as species (more
simultaneous infection by particles containing all of the dif-
than 30,000 strains of viruses exist in collections around
ferent genome segments. Although this does not seem to pose
the world), and classified these 5450 species into 287 genera
a problem for the transmission of plant viruses, it must pose a
belonging to 73 families plus a number of "floating" genera that
problem for the transmission of animal viruses since such
have not yet been assigned to a family. An overview of
animal viruses have not been found. This difference probably
these families, in which viruses that cause human disease
arises because of different modes of transmission, the fact
are emphasized, is shown in Table 1.2. Included in the table
that many plant viruses grow to exceptionally high titers, and
is the type of nucleic acid that serves as the genome, the
the fact that many plants grow to very high density.
genome size, the names of many families, and the major
groups of hosts infected by viruses within each grouping.
For many families the names and detailed characteristics are
The ICTV Classification of Viruses
not shown here, but a complete listing of families can be
The International Committee on Taxonomy of Viruses
found in the reports of the ICTV on virus taxonomy or in
The Encyclopedia of Virology (2nd ed.). Tables that describe
(ICTV), a committee organized by the Virology Division
of the International Union of Microbiological Societies, is
the members of families that infect humans are presented in
attempting to devise a uniform system for the classification
the chapters that follow in which the various virus families
and nomenclature of all viruses. Viruses are classified into
are considered in some detail.
species on the basis of a close relationship. The decision as
to what constitutes a species is arbitrary because a species
usually contains many different strains that may differ sig-
AN OVERVIEW OF THE REPLICATION CYCLE
nificantly (10% or more) in nucleotide sequence. Whether
OF VIRUSES
two isolates should be considered as being the same spe-
cies rather than representing two different species can be
Receptors for Virus Entry
controversial. Virus species that exhibit close relationships
The infection cycle of an animal virus begins with its
are then grouped into a genus. Species within a genus usu-
attachment to a receptor expressed on the surface of a sus-
ally share significant nucleotide sequence identity demon-
ceptible cell, followed by penetration of the genome, either
strated by antigenic cross-reaction or by direct sequencing
TABLE 1.2
Major Virus Families
Major hosts (number of members infecting that host)a
Nucleic acid
Genome size
Segments
Family
Genera
Vertebrates (35 + 9T)b, insects (27), plus 15 Uc
DS DNA
130375 kbp
1
Poxviridae
8+3
170190 kbp
1
Asfariviridae
1
Vertebrates (1)
170400 kbp
1
Iridoviridae
3+2
Vertebrates (2 + 5T), insects (6 + 11T)
120220 kbp
1
Herpesviridae
9
Vertebrates (61 + 7T + 56U)
80180 kbp
1
Baculoviridae
2
Insects (36 + 8T)
2848 kbp
1
Adenoviridae
4
Vertebrates (32 + 9T)
5 kbp
1
Polyomaviridae
1
Vertebrates (12)
6.88.4 kbp
1
Papillomaviridae
16
Vertebrates (7 + 88T + 13U)
Various
1
Several families
--
Bacteria (42 + 368T)
SS DNA
46 kbp
1
Parvoviridae
5+4
Vertebrates (33 + 3T), insects (6 + 18T)
Various
1
Several families
--
Bacteria (43 + 38T), plants (98 + 11T)
DS RNA
2030 kbp
1012
Reoviridae
6+2+4
Vertebrates (52 + 24T), insects (1 + 7T), plants (13 + 1T)
5.9 kbp
2
Birnaviridae
2+1
Vertebrates (3), insects (1)
4.67.0 kbp
1 or 2
Three families
--
Fungi (7 + 7T), plants (30 + 15T), protozoans (14)
SS (+)RNA
2833 kb
1
Coronaviridae
2
Vertebrates (17 + 1T)
1316 kb
1
Arteriviridae
1
Vertebrates (4)
1013 kb
1
Togaviridae
2
Vertebrates (insect vectors)(28)
1012 kb
1
Flaviviridae
3
Vertebrates (some insect vectors) (59 + 4T)
78.5 kb
1
Picornaviridae
9
Vertebrates (30 + 1T + 23U)
78 kb
1
Astroviridae
2
Vertebrates (9)
8 kb
1
Caliciviridae
4
Vertebrates (6 + 1T)
7.2 kb
1
Hepeviridae
1
Vertebrates (1)
Various
1 to 3
Many families
--
Plants (496 + 84T + 5U)
SS (-)RNA
1516 kb
1
Paramyxoviridae
7
Vertebrates (34 + 2U)
19 kb
1
Filoviridae
2
Vertebrates (5)
1116
1
Rhabdoviridae
4+2
Vertebrates (23 + 25T + 40U), invertebrates (20U), plants (15)
6 kb
1
Bornaviridae
1
Vertebrates (1)
1015 kb
8
Orthomyxoviridae
5
Vertebrates (7)
1223 kb
3
Bunyaviridae
4+1
Vertebrates and insect vectors (86 + 20T), plants (9 + 7T)
11 kb
2
Arenaviridae
1
Vertebrates (19 + 1T)
SS RNA RT
710 kb
dimer
Retroviridae
7
Vertebrates (53 + 2T)
DNA Intermediate
DS DNA RT
3 kb
1
Hepadnaviridae
2
Vertebrates (5 + 1T)
RNA intermediate
8 kb
1
Caulimoviridae
6
Plants (26 + 10T)
RNA intermediate
a
Vertebrates in red indicate humans are among the vertebrates infected. Vertebrates in blue indicate non-human hosts only; plant hosts are in green; insect
hosts in yellow; bacterial hosts are black.
b
T = tentatively assigned to a particular genus.
c
U = assigned to the family, but not to any particular genus within the family.
Source: Data for this table is from Fauquet et al. (2005).
naked or complexed with protein, into the cytoplasm.
sory receptor is not required for virus entry even where used,
Binding often occurs in several steps. For many viruses, the
but such binding does accelerate the rate of binding and
virion first binds to an accessory receptor that is present in
uptake of the virus.
high concentration on the surface of the cell. These accessory
Binding to a high-affinity, virus-specific receptor is
receptors are usually bound with low affinity and binding
required for virus entry, and virus may be transferred to its
often has a large electrostatic component. Use of accessory
high-affinity receptor after primary binding to an accessory
receptors seems to be fairly common among viruses adapted
receptor, or may bind directly to its high-affinity receptor.
to grow in cell culture but less common in primary isolates
Cells that fail to express the appropriate receptor cannot be
of viruses from animals. This first stage binding to an acces-
infected by the virus. These receptors are specifically bound
by one or more of the external proteins of a virus. Each virus
disparate receptors. Fig. 1.4 illustrates a number of receptors
uses a specific receptor (or perhaps a specific set of recep-
used by different retroviruses (family Retroviridae). These
tors) expressed on the cell surface, and both protein recep-
receptors differ widely in their structures and in their cellular
tors and carbohydrate receptors are known. In some cases,
functions. Where known, the region of the cellular receptor
unrelated viruses make use of identical receptors. A protein
that is bound by the virus is indicated. Table 1.3 lists recep-
called CAR (Coxsackie-adenovirus receptor), a member of
tors used by different herpesviruses (Herpesviridae) and dif-
the immunoglobulin (Ig) superfamily, is used by the RNA
ferent coronaviruses.
virus Coxsackie B virus (Picornaviridae) and by many ade-
In addition to the requirement for a high-affinity or pri-
noviruses (Adenoviridae), which are DNA viruses. Sialic
mary receptor, many viruses also require a coreceptor in
acid, a carbohydrate attached to most glycoproteins, is used
order to penetrate into the cell. In the current model for
by influenza virus (family Orthomyxoviridae), human coro-
virus entry, a virus first binds to the primary receptor and
navirus OC3 (family Coronaviridae), reovirus (Reoviridae),
then binds to the coreceptor. Only on binding to the core-
bovine parvovirus (Parvoviridae), and many other viruses.
ceptor can the virus enter the cell. The best studied example
Conversely, members of the same viral family may use widely
is HIV, which uses the cell surface molecule called CD4 as
A. Gammaretrovirus Receptors
Host cell
plasma
membrane
C
C
N
N
N
C
Virus
MoMLV
GALV/FeLV
MLV
Amphotropic
Ecotropic
Type
hPiT-1
rPiT-2
mCAT-1
Protein
679
652
622
# amino acids
Phosphate transporter
Similar to hPiT-1
Basic amino acid
Protein type
transporter
N
C
B. Other Retrovirus Receptors
N
N
Host cell
plasma
membrane
C
N
C
C
BLV
ALV (subgroup A)
Virus
HIV
Deltaretrovirus
Genus
Alpharetrovirus
Lentivirus
BLV receptor
Protein
Tv-a
CD4 + CXCR4 (or CCR5)
730
# amino acids
369
458 + 353
Unknown
Protein type
LDL-receptor-like
Immunoglobulin and
chemokine receptor
Region to which env
Transmembrane domains
Disulfide bridges
proteins bind
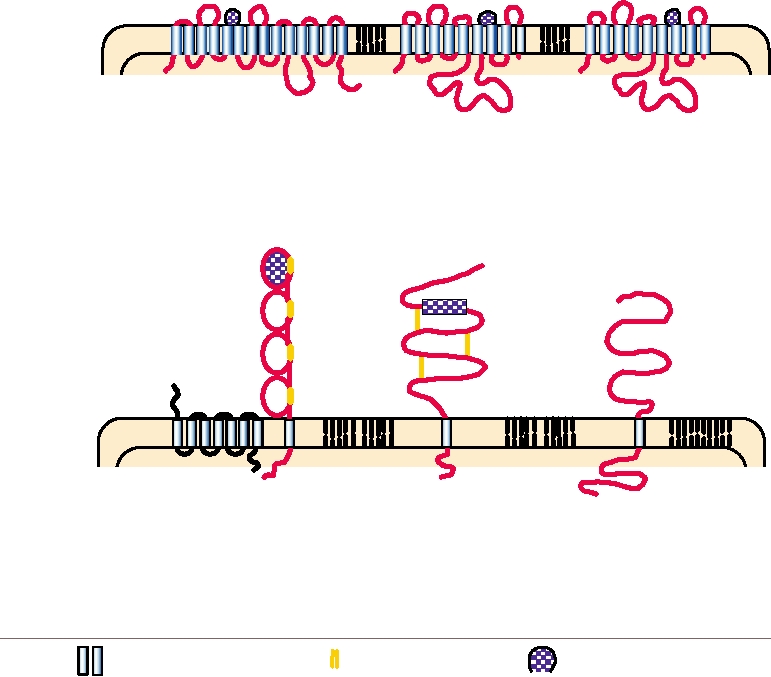
FIGURE 1.4
Cellular receptors for retroviruses. The structures of various retrovirus receptors are shown schematically
to illustrate their orientation in the cell plasma membrane. The receptors for the gammaretroviruses contain multiple
transmembrane domains and have known cellular functions. The HIV receptor consists of a molecule of CD4 plus a
chemokine receptor such as CXCR4. The receptor for alpharetroviruses is a Type II membrane protein similar to the
LDL receptor, with the N terminus in the cytoplasm. Little is known about the cellular function of the BLV receptor,
other than its orientation as a Type I membrane protein. Abbreviations: MLV, murine leukemia virus; GALV, gibbon/ape
leukemia virus; FeLV, feline leukemia virus; HIV, human immunodeficiency virus; ALV, avian leukosis virus; BLV,
bovine leukemia virus; LDL, low density lipoprotein. Adapted from Fields et al. (1996) p. 1788 and Coffin et al. (1997)
pp. 7682.
TABLE 1.3 Viruses Within a Family that Use Unrelated Receptors
Family
Virus
High-affinity receptor
Accessory receptor
Herpesviridae
Alpha
Herpes simplex
HIgR (CD155 family)
Heparan sulfate
HVEM (TNF receptor family)
Pseudorabies
140 kD heparan sulfate proteoglycan
85 kD integral membrane protein
CD155 and related proteins
Beta
Cytomegalovirus
protein?? unidentified
Heparan sulfate
Gamma
Bovine herpesvirus
56 kD protein
Heparan sulfate
Epstein Barr
CD21 (CR2 receptor)
Coronaviridae
Group 1
TGEVa
Porcine APN: aminopeptidase N
Porcine
FIPVa
Feline
Feline APN: aminopeptidase N
Human
229e
Human APN: human aminopeptidase N
Human
NL63
ACE2: Human angiotensin-converting enzyme 2
Group 2
Human
SARS
ACE2: Human angiotensin-converting enzyme 2
Murine
Mouse hepatitis
CEACAM1: Carcinoembryonic antigen-related cell
adhesion molecule 1b
Bovine
Bovine coronavirus
Sialic acid residues on glycoproteins and glycolipids
a
Virus abbreviation: TGEV, transmissible gastroenteritis virus (of swine); FIPV, feline infectious peritonitis virus.
b
Note that entry of mouse hepatitis variants of extended host range is independent of CEACAM1, and instead uses heparan sulfate as an entry receptor.
a primary receptor and various chemokines as coreceptors
Other surface proteins used as receptors include the
vibronectin receptor αvβ3, used by several members of the
(see later).
The nature of the receptors utilized by a virus determines
Picornaviridae; aminopeptidase N, used by some corona-
in part its host range, tissue tropism, and the pathology of the
viruses; CD55, used by Coxsackie A21 virus; the different
disease caused by it. Thus, the identification of virus recep-
proteins illustrated in Fig. 1.4; and other proteins too numer-
tors is important, but identification of receptors is not always
ous to describe here. The receptors used by four viruses
straightforward.
are described in more detail as examples of the approaches
used to identify receptors and their importance for virus
pathology.
Primary (High-Affinity) Receptors
One well-characterized receptor is that for poliovirus,
Many members of the Ig superfamily are used by viruses
which attaches to a cell surface molecule that is a member
as high-affinity receptors, as illustrated in Fig. 1.5. The Ig
of the Ig superfamily (Fig. 1.5). The normal cellular func-
superfamily contains thousands of members, which play
tion of this protein is unknown. It was first called simply
important roles in vertebrate biology. The best known mem-
the poliovirus receptor or PVR, but has now been renamed
bers are found in the immune system (Chapter 10), from
CD155, following a scheme for the designation of cell sur-
which the family gets its name. Members of this superfamily
face proteins. Poliovirus will bind only to the version of this
contain one or more Ig domains of about 100 amino acids that
molecule that is expressed in primates, and not to the version
arose by duplication of a prototypical gene. During evolu-
expressed in rodents, for example. Thus, in nature, polio-
tion of the superfamily, thousands of different proteins arose
virus infection is restricted to primates. Although chicken
by a combination of continuing gene duplication, sequence
cells or most mammalian cells that lack CD155 are resistant
divergence, and recombination. Many proteins belonging to
to poliovirus infection, they can be transfected with the viral
this superfamily are expressed on the surface of cells, where
RNA by a process that bypasses the receptors. When infected
they serve many functions, and many have been usurped by
in this way, they produce a full yield of virus, showing that
animal viruses for use as receptors.
the block to replication is at the level of entry.
HLA
Carcinoembryonic CD4
CD155
ICAM-1
HIgR
Receptor
antigens
Virus
Semliki
mouse
HSV1, HSV2
HIV-1
poliovirus rhinovirus
bovine herpesvirus Forest
hepatitis
Togaviridae
Coronaviridae
Retroviridae
Picornaviridae
Herpesviridae
Family
dsDNA-large
ssRNA
ssRNA
RNA/RT
ssRNA-small
Genome
enveloped
enveloped
nonenveloped
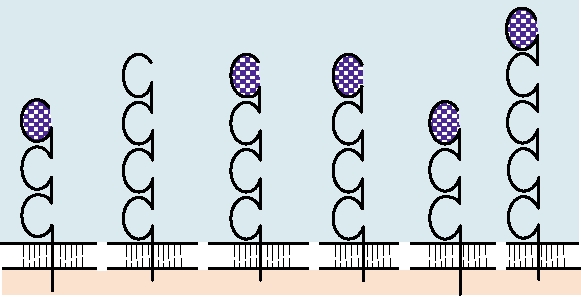
FIGURE 1.5 Diagrammatic representation of immunoglobulin superfamily membrane proteins that are used as
receptors by viruses. The domains indicated by cross-hatching have been shown to be required for receptor activity.
ssRNA, single-strand RNA; dsDNA, double-strand DNA; RNA/RT, RNA reverse transcribed into DNA.
Cells lacking CD155 have been transfected with expres-
be faithfully reproduced. Although these transgenic mice can
sion clones so that they express CD155, and these modified
be infected only by injection of virus and not by ingestion,
cells are sensitive to infection with poliovirus. This was, in
the normal route of poliovirus infection in humans, a small
fact, the way the receptor was identified. Such a system also
animal model for poliomyelitis is valuable for the study of
allows the testing of chimeric receptors, in which various
virus pathology or for vaccine development. To date, our
domains of CD155 come from the human protein and other
information on the pathology of poliovirus in the CNS was
parts come from the homologous mouse protein, or even
obtained only from experimental infection of nonhuman
from entirely different proteins like CD4. In this way it was
primates, which are very expensive to maintain, or from
shown that only the distal Ig domain from human CD155
humans naturally infected with the virus.
(cross-hatched in Fig. 1.5) is required for a chimeric protein
As a second example of virusreceptor interactions, HIV
to function as a receptor for poliovirus.
utilizes as its receptor a cell surface molecule known as CD4,
In humans, CD155 is expressed on many cells, including
which is also a member of the Ig superfamily (Figs 1.4 and
cells of the gut, nasopharynx, and the central nervous system
1.5). As described later, a coreceptor is also required. CD4
(CNS). Infection begins in the tonsils, lymph nodes of the
is primarily expressed on the surface of certain lymphocytes
neck, Peyer's patches, and the small intestine. In more than
(described in more detail in Chapter 10). Furthermore, the
98% of cases, the infection progresses no further and no ill-
virus has a narrow host range and will bind with high effi-
ness, or only minor illness, results. In some cases, however,
ciency only to the human version of CD4 (Fig. 1.4). Thus,
virus spreads to the CNS, probably both by passing through
humans are the primary host of HIV. Immune function
is impaired over time as helper CD4+ T cells, which are
the bloodbrain barrier and through retrograde axonal trans-
port. Once in the CNS, the virus expresses an astounding
required for an immune response directed against infectious
preference for motor neurons, whose destruction leads to
agents, are killed by virus infection, leading to the observed
paralysis or even death via a disease called poliomyelitis.
syndrome of AIDS (acquired immunodeficiency syndrome).
This preference for motor neurons, and the failure of the
The virus can also infect cells of the monocyte-macrophage
virus to grow in other tissues, is not understood. Athough
lineage, and possibly other cells in the CNS, leading to
CD155 is required for virus entry, other factors within the
neurological manifestations.
cell are also important for efficient virus replication.
As a third example of virusreceptor interaction, among
Making use of the CD155 gene, transgenic mice have
the receptors used by Sindbis virus (family Togaviridae)
been generated in which the syndrome of poliomyelitis can
is the high-affinity laminin receptor. Sindbis virus is an
arbovirus, that is, it is arthropod-borne. In nature it alternates
proteins, one called HIgR (for herpesvirus Ig-like receptor)
between replication in mosquitoes, which acquire the virus
and the other called either PRR-1 (for poliovirus receptor
when they take a blood meal from an infected vertebrate, and
related) or HveA (for herpesvirus entry mediator A), appear
higher vertebrates, which acquire the virus when bitten by
to be splice variants that have the same ectodomain.
an infected mosquito. The high-affinity laminin receptor is a
Heparan sulfate may serve as an accessory receptor for
cell adhesion molecule that binds to laminin present in base-
the other viruses shown in Table 1.4, or it may serve as a pri-
ment membranes. It has been very highly conserved during
mary receptor for some or all. It was thought that it may be
evolution, and Sindbis virus will bind to both the mosquito
a primary receptor for dengue virus (family Flaviviridae),
version and the mammalian version of this protein. Viruses
but recent work has identified other candidates as the pri-
with broad host ranges, such as arboviruses, must use recep-
mary receptor. In the case of Sindbis virus, the situation is
tors that are highly conserved, or must have evolved the
complex and interesting. Primary isolates of the virus do
ability to use different receptors in different hosts.
not bind to heparan sulfate. Passage of the virus in cultured
Finally, as a fourth example, the receptor for influenza
cells selects for viruses that do bind to heparan sulfate, and
virus (family Orthomyxoviridae) is sialic acid covalently
which infect cultured cells more efficiently. It is thought
linked to glycoproteins or glycolipids present at the cell sur-
that selection for heparin sulfate binding upon passage of
face. Because sialic acid is expressed on many different cells
the virus in the laboratory speeds up the process of infec-
and in many different organisms, the virus has the potential to
tion in cultured cells because virus bound to the cell surface
have a very wide host range. The virus infects many birds and
by binding to heparin sulfate can diffuse in two dimensions
mammals, and its maintenance in nature depends on its ability
rather than three to encounter its high-affinity receptor. In
to infect such a broad spectrum of animals. The epidemiology
infected animals, however, heparin sulfate binding attenu-
of influenza virus will be considered in Chapter 4.
ates the virus, perhaps allowing the animal to clear the virus
more quickly.
Many viruses absolutely require a coreceptor for entry,
Accessory Receptors and Coreceptors
in addition to the primary receptor to which the virus first
binds. The best studied example is HIV, which requires one
The process by which a virus binds to a cell and pen-
of a number of chemokine receptors as a coreceptor. Thus a
etrates into the cytoplasm may be complicated by the par-
ticipation of more than one cellular protein in the process.
Some viruses may be able to use more than one primary
TABLE 1.4 Viruses That Bind to Heparin-Like
receptor, which thus serve as alternative receptors. Second,
Glycosaminoglycans
many viruses appear to first bind to a low-affinity receptor
or accessory receptor before transfer to a high-affinity recep-
Virus
Family
High affinity receptor
tor by which the virus enters the cell. Third, many viruses
absolutely require a coreceptor, in addition to the primary
RNA viruses
Sindbis
Togaviridae
High affinity laminin
receptor, for entry.
receptor
Many viruses, belonging to different families, have been
Dengue
Flaviviridae
???
shown to bind to glycosoaminoglycans such as heparan sul-
Hepatitis C
Flaviviridae
CD81
fate (Table 1.4), which are expressed on the surface of many
αvβ3 integrin
Foot and mouth disease
Picornaviridae
cells. In at least some cases, however, such as for human her-
pes simplex virus (HSV) (family Herpesviridae), heparan
Respiratory syncytial
Paramyxoviridae
???
sulfate is not absolutely required for the entry of the virus.
Retroviruses
Cells that do not express heparan sulfate or from which it has
HIV-1
Retroviridae
CD4 (Ig superfamily)
been removed can still be infected by HSV. Heparan sulfate
does dramatically increase the efficiency of infection, how-
DNA viruses
ever. The current model is that HSV first binds to heparan
Vaccinia
Poxviridae
EGF receptor ???
sulfate with low affinity and is then transferred to the pri-
Syndecan-1a
Human papillomavirus
Papillomaviridae
mary receptor for entry. In this model, heparan sulfate serves
Herpes simplex
Herpesviridae
HIgR (CD155 family)
an accessory function, which can be dispensed with.
Adeno-associated type 2 Parvoviridae
FGFR1
The primary receptor for HSV has now been identified
as a protein belonging to the Ig superfamily (Fig. 1.5). This
a
In this case the heparan sulfate proteoglycan appears to be the primary
protein is closely related to CD155, and, in fact, CD155 will
receptor protein.
serve as a receptor for some herpesviruses, but not for HSV.
Abbreviations used: EGF receptor, epidermal growth factor receptor;
The story is further complicated by the fact that more than
HIgR, herpes immunoglobulin-like receptor; CD155, the poliovirus
one protein can serve as a receptor for HSV. Two of these
receptor; FGFR1, human fibroblast growth factor receptor 1.
mouse cell that is genetically engineered to express human
by insect or fungal pests, in fact, with which the virus has a
CD4 will bind HIV, but binding does not lead to entry of the
specific association. There remains the possibility that spe-
virus into the cell. Only if the cell is engineered to express
cific receptors will be identified in the future, however, for
both human CD4 and a human chemokine receptor can the
at least some plant viruses.
virus both bind to and enter into the cell. It is thought that
binding to the first or primary receptor induces conforma-
Penetration
tional changes in the virion that allow it to bind to the second
or coreceptor.
After the virus binds to a receptor, the next step toward
The requirement for a coreceptor has important impli-
successful infection is the introduction of the viral genome
cations for the pathology of HIV. Chemokines are small
into the cytoplasm of the cell. In some cases, a subviral par-
proteins, secreted by certain cells of the immune system, that
ticle containing the viral nucleic acid is introduced into the
serve as chemoattractants for lymphocytes. They are impor-
cell. This particle may be the nucleocapsid of the virus or
tant regulators of the immune system and are described in
it may be an activated core particle. For other viruses, only
Chapter 10. Different classes of lymphocytes express recep-
the nucleic acid is introduced. The protein(s) that promotes
tors for different chemokines at their surface. To simplify the
entry may be the same as the protein(s) that binds to the
story, macrophage-tropic (M-tropic) strains of HIV, which is
receptor, or it may be a different protein in the virion.
the virus most commonly transmitted sexually to previously
For enveloped viruses, penetration into the cytoplasm
uninfected individuals, require a coreceptor called CCR5 (a
involves the fusion of the envelope of the virus with a cel-
receptor for β chemokines). Human genetics has shown that
lular membrane, which may be either the plasma membrane
two mutations can block the expression of CCR5. One is a
or an intracellular membrane. Fusion is promoted by a
32-nucleotide deletion in the gene, the second is a mutation
fusion domain that resides in one of the viral surface pro-
that results in a stop codon in the CCR5 open reading frame
teins. Activation of the fusion process is thought to require a
(ORF). The deletion mutation is fairly common, present in
change in the structure of the viral fusion protein that exposes
about 20% of Caucasians of European descent, whereas the
the fusion domain. For viruses that fuse at the plasma mem-
stop codon mutation has been reported in only one indi-
brane, interaction with the receptor appears to be sufficient
vidual. Individuals who lack functional CCR5 because they
to activate the fusion protein. In the case of viruses that
are homozygous for the deleted form, or in the case of one
fuse with intracellular membranes, the virus is internalized
individual, heterozygous for the deletion but whose second
via various cellular vesicular pathways, which may differ
copy of CCR5 has the stop codon, are resistant to infection
depending upon the virus. The best studied internalization
by HIV. Heterozygous individuals who have only one func-
process is endocytosis into clathrin-coated vesicles and pro-
tional copy of the CCR5 gene appear to be partially resist-
gression through the endosomal pathway. During transit,
ant. Although they can be infected with HIV, the probability
the clathrin coat is lost and the endosomes become progres-
of transmission has been reported to be lower, and once
sively acidified. On exposure to a defined acidic pH (often
infected, progression to AIDS is slower. During the course
~56), activation of the fusion protein occurs and results in
of infection by HIV, T-cell-tropic strains (T-tropic) of HIV
fusion of the viral envelope with that of the endosome. In
arise that require a different coreceptor, called CXCR4 (a
either case, the nucleocapsid of the virus is present in the
receptor for α chemokines). After the appearance of T-tropic
cytoplasm after fusion.
virus, both M-tropic and T-tropic strains cocirculate. The
A dramatic conformational rearrangement of the hemag-
requirement for a new coreceptor is associated with muta-
glutinin glycoprotein (HA) of influenza virus, a virus that
tions in the surface glycoprotein of HIV. The presence of
fuses with internal membranes, has been observed by X-ray
T-tropic viruses is associated with more rapid progression to
crystallography of HA following its exposure to low pH. HA,
severe clinical disease.
which is cleaved into two disulfide-bonded fragments HA1
and HA2, forms trimers that are present in a spike on the sur-
face of the virion. The atomic structure of an HA monomer
Entry of Plant Viruses
is illustrated in Fig. 1.6. HA1 (shown in blue) is external and
Many plant viruses are important pathogens of food crops
derived from the N-terminal part of the precursor. It contains
and have been intensively studied. No specific receptors
the domain (indicated with a star in the figure) that binds to
have been identified to date, and it has been suggested that
sialic acid receptors. HA2 (shown in red) is derived from the
virus penetration of plant cells requires mechanical damage
C-terminal part of the precursor and has a C-terminal anchor
to the cell in order to allow the virus entry. Such mechanical
that spans the viral membrane. The fusion domain (yellow)
damage can be caused by farm implements or by damage
is present at the N terminus of HA2, hidden in a hydrophobic
to the plant caused by insects such as aphids or leafhoppers
pocket within the spike near the lipid bilayer of the virus enve-
that feed on the plants. Many plant viruses are transmitted
lope. Exposure to low pH results in a dramatic rearrangement
A.
S -S
S
TM
N
C
HA1 (328aa)
16aa
HA2 (221aa)
B9.
C9.
B.
C.
40
40
A
B
153
G
76
76
76
B
153
129
E
C
F
D
C
105
105
A
105
105
D
1(N)
38
40
153
F
G
E
153
H
129
1(N)
175
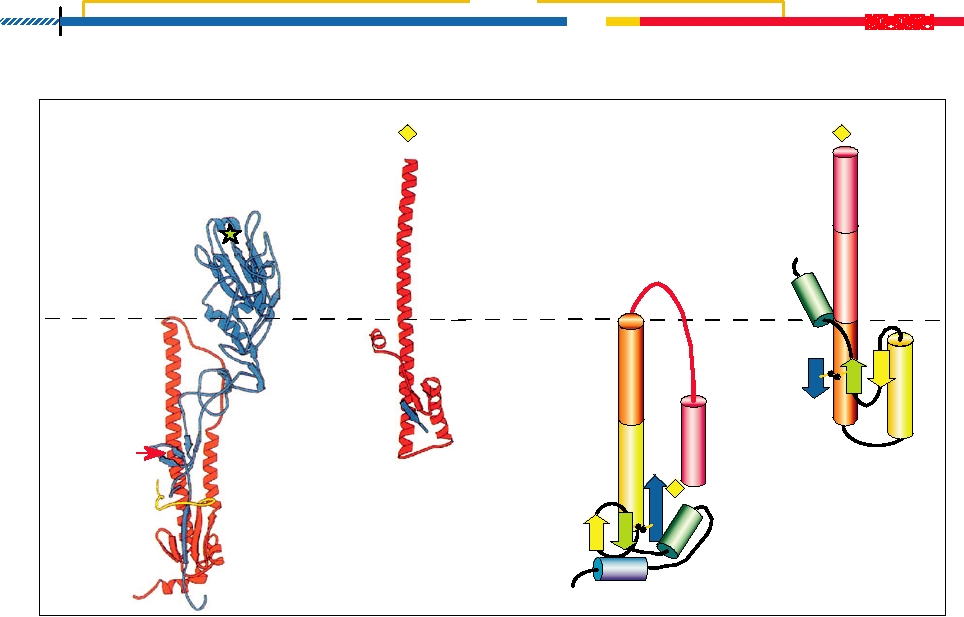
FIGURE 1.6 The folded structure of the influenza hemagglutinin and its rearrangement when exposed to low pH. (A) A
schematic of the cleaved HA molecule. S is the signal peptide, TM is the membrane-spanning domain. HA1 is in blue, HA2
is in red, and the fusion peptide is shown in yellow. The same color scheme is used in (B) and (C). (B) X-ray crystallographic
structure of the HA monomer. TM was removed by proteolytic digestion prior to crystallization. The receptor-binding
pocket in HA1 is shown with a green star. In the virion HA occurs as a trimeric spike. (C) The HA2 monomer in the fusion
active form. The fragment shown is produced by digesting with thermolysin, which removes most of HA1 and the fusion
peptide of HA2. Certain residues are numbered to facilitate comparison of the two forms. The approximate location of the
fusion peptide before thermolysin digestion is indicated with a yellow diamond. (B′) Diagrammatic representation of the
HA2 shown in (B), with α helices shown as cylinders and β sheets as arrows. The disulfide link between HA1 and HA2 is
shown in ochre. The domains of HA2 are color coded from N terminus to C terminus with a rainbow. (C′) Diagrammatic
representation of the fusion-active form shown in (C). Redrawn from Fields et al. (1996) p. 1361, with permission.
of HA that exposes the hydrophobic peptide and transports
rearrange to form a hexameric helical bundle, which forces
it more than 100 Å upward, where it is thought to insert into
the cellular membrane and the viral membrane together,
the cellular membrane and promote fusion. It is assumed that
resulting in fusion. Fusion can be blocked by peptides that
similar events occur for all enveloped viruses, whether fusion
bind to one or the other of the trimeric bundles, preventing
is at the cell surface or with an internal membrane.
the formation of the hexameric bundle.
Studies with HIV have further refined our understand-
For nonenveloped viruses, the mechanism by which the
ing of the fusion process. The external glycoproteins of HIV
virus breaches the cell membrane is less clear. After binding
are also synthesized as a precursor that is cleaved into an
to a receptor, somehow the virus or some subviral compo-
N-terminal protein (called gp120) and a C-terminal, mem-
nent ends up on the cytoplasmic side of a cellular membrane,
brane-spanning protein (called gp41). Like the case for influ-
the plasma membrane for some viruses, or the membrane of
enza (and many other enveloped viruses), the glycoproteins
an endosomal vesicle for others. It is believed that the inter-
form trimers. A model for the process of fusion is shown
action of the virus with a receptor, perhaps potentiated by
in Fig. 1.7. The external gp120 binds to the receptor CD4
the low pH in endosomes for those viruses that enter via the
and then to the coreceptor chemokine. The fusion domain
endosomal pathway, causes conformational rearrangements
at the N terminus of gp41 rearranges and penetrates the host
in the proteins of the virus capsid that result in the forma-
cell membrane. Two trimeric helical bundles in gp41 then
tion of a pore in the membrane. In the case of poliovirus, it
C
Co-receptor C
gp120
Virus interacts with
CD4
host-cell receptors
Viral membrane
Exposed
Viral prehairpin
trimeric
intermediate forms
N-peptide
gp41
Fully
exposed
C-peptide
Myc
+ 5-Helix/3Myc
+ C-peptides
Hairpin
gp41
gp41
Inhibited
Inhibited
Myc
intermediate
intermediate
FUSION
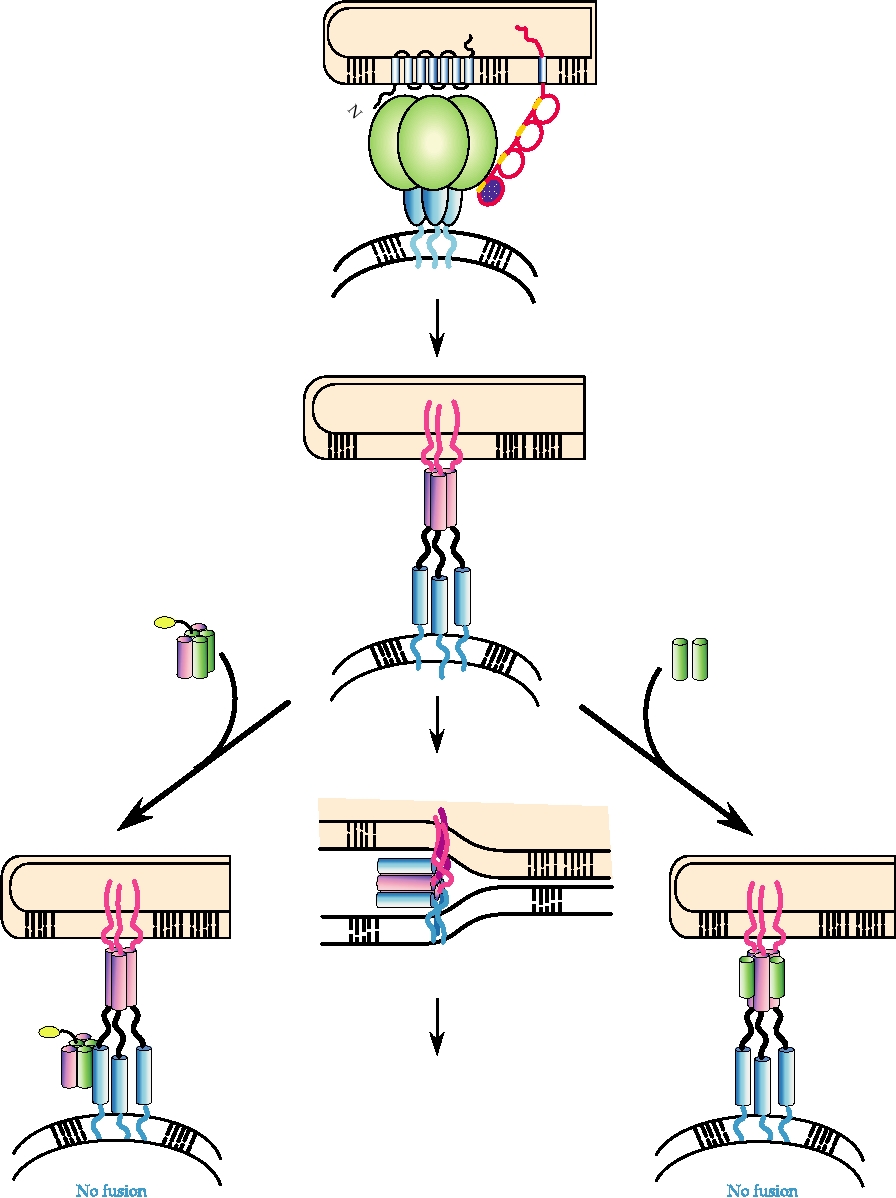
FIGURE 1.7
A model for HIV-1 membrane fusion and two forms of inhibition. In the native state gp120 partially shields
gp41. When gp120 interacts with receptors and coreceptors on the host cell surface, gp41 undergoes a configurational
rearrangement to the transient prehairpin intermediate, in which both the N and C peptides of gp41 are exposed. Fusion
can be inhibited either by binding of C peptides to the trimeric N-peptide bundle or by binding of 5-helix/3Myc to a gp41
C peptide. Figure is adapted from Figure 6 in Koshiba and Chan (2003).
Replication and Expression of the
is known that interactions with receptors in vitro will lead
to conformational rearrangements of the virion that result
Virus Genome
in the release of one of the virion proteins, called VP4. The
The replication strategy of a virus, that is, how the
N terminus of VP4 is myristylated and thus hydrophobic
genome is organized and how it is expressed so as to lead to
[myristic acid = CH3(CH2)12COOH]. It is proposed that the
the formation of progeny virions, is an essential component
conformational changes induced by receptor binding result
in the classification of a virus. Moreover, it is necessary to
in the insertion of the myristic acid on VP4 into the cell
understand the replication strategy in order to decipher the
membrane and the formation of a channel through which
pathogenic mechanisms of a virus and, therefore, to design
the RNA can enter the cell. It is presumed that other viruses
strategies to interfere with viral disease.
also have hydrophobic domains that allow them to enter. A
number of other viruses also have a structural protein with a
DNA Viruses
myristilated N terminus that might promote entry. In some
viruses, there is thought to be a hydrophobic fusion domain
A simple schematic representation of the replication of a
in a structural protein that provides this function.
DNA virus is shown in Fig. 1.8. After binding to a receptor
The entry process may be very efficient. In the case of
and penetration of the genome into the cell, the first event in
enveloped viruses, there is evidence that at least for some
viruses the specific infectivity in cultured cells can be one
(all virions can initiate infection), and successful penetration
is thought to be efficient for all enveloped viruses. For non-
enveloped viruses, the situation varies. The specific infectiv-
ity of reoviruses assayed in cultured cells can be almost one
Uncoating
but for other viruses, entry may be less efficient. For exam-
ple, the specific infectivity of poliovirus in cultured cells is
Genome (DNA)
usually less than 1%. In general it is not known how such
specific infectivites assayed in cultured cells relate to the
infectivity of the virus when infecting host animals.
During entry of at least some viruses it is known that cel-
Transcription
lular functions must be activated and it is thought that bind-
+
mRNAs
ing of the virus to its receptor signals the cell to do something
that is required for virus penetration. For example, binding
Translation
of adenoviruses activates a pathway that results in polym-
Modification
+
erization of actin and endocytosis of the virus. As a second
DNA replication
example, internalization of the polyomavirus SV40 is regu-
+
lated by at least five different kinases. These activations of
Assembly
cellular pathways are only beginning to be unraveled.
Following initial penetration into the cytoplasm, further
uncoating steps must often occur. It has been suggested that,
NUCLEUS
Release
at least in some cases, translation of the genomic RNA of
plus-strand RNA viruses may promote its release from the
EUKARYOTIC HOST CELL
nucleocapsid. In other words, the ribosomes may pull the
RNA into the cytoplasm. In other cases, specific factors in
Host RNA polymerase
the host cell, or the translation products of early viral tran-
scripts, have been proposed to play a role in further uncoat-
Host DNA polymerase
ing.
Viral-encoded factor
It is interesting to note that bacteriophage face the prob-
lem of penetrating a rigid bacterial cell wall, rather than
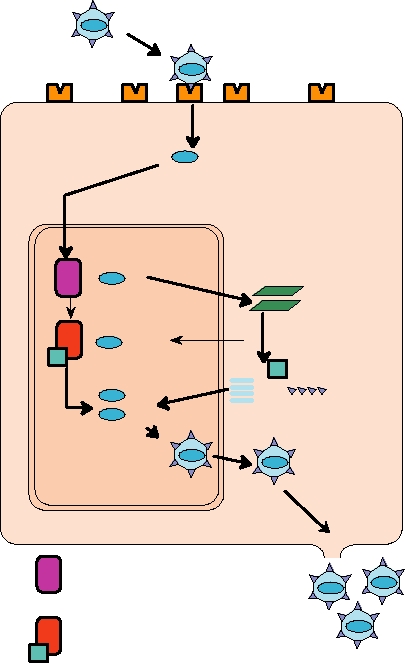
FIGURE 1.8
General replication scheme for a DNA virus. After
one of simply penetrating a plasma membrane or intracel-
a DNA virus attaches to a cellular membrane receptor, the virus DNA
lular membrane. Many bacteriophage have evolved a tail
enters the cell and is transported to the cell nucleus. There it is transcribed
into mRNA by host RNA polymerase. Viral mRNAs are translated by
by which they attach to the cell surface, drill a hole into
host ribosomes in the cytoplasm, and newly synthesized viral proteins,
the cell, and deliver the DNA into the bacterium. In some
both structural and nonstructural, are transported back to the nucleus.
phage, the tail is contractile, leading to the analogy that the
After the DNA genome is replicated in the nucleus, either by the host
DNA is injected into the bacterium. Tailless phage are also
DNA polymerase or by a new viral-encoded polymerase, progeny virus
known that introduce their DNA into the bacterium by other
particles are assembled and ultimately released from the cell. Adapted
from Mims et al. (1993) p. 2.3.
mechamisms.
the replication of a DNA virus is the production of mRNAs
by DNA polymerase. Unit sized genomes are cut from the
from the viral DNA. For all animal DNA viruses except pox-
multilength genomes that result from this replication scheme
viruses, the infecting genome is transported to the nucleus
and are packaged into virions.
where it is transcribed by cellular RNA polymerase. The pox-
Once initiated, the progression of the replication fork is dif-
viruses replicate in the cytoplasm and do not have access to
ferent in different viruses, as illustrated in Fig. 1.9. In SV40
host cell polymerases. Therefore, in poxviruses, early mRNA
(family Polyomaviridae), for example, the genome is circular.
is transcribed from the incoming genome by a virus-encoded
An RNA primer is synthesized by primase to initiate replica-
RNA polymerase that is present in the virus core. For all ani-
tion, and the replication fork then proceeds in both directions.
mal DNA viruses, translation of early mRNA is required for
The product is two double-strand circles. In the herpesviruses,
viral DNA replication to proceed. Early gene products may
the genome is circular while it is replicating but the replication
include DNA polymerases, proteins that bind to the origin of
fork proceeds in only one direction. A linear double-strand
replication and lead to initiation of DNA replication, proteins
DNA is produced by what has been called a rolling circle.
that stimulate the cell to enter S phase and thus increase the
For this, one strand is nicked by an endonuclease and used
supply of materials required for DNA synthesis, or products
as a primer. The strand displaced by the synthesis of the new
required for further disassembly of subviral particles.
strand is made double stranded by the same mechanism used
The initiation of the replication of a viral genome is a
by the host cell for lagging strand synthesis. In adenoviruses,
specific event that requires an origin of replication, a spe-
in contrast, the genome is linear and the replication fork pro-
cific sequence element that is bound by cellular and (usu-
ceeds in only one direction. A single-strand DNA is displaced
ally) viral factors. Once initiated, DNA replication proceeds,
during the progression of the fork and coated with viral pro-
catalyzed by either a cellular or a viral DNA polymerase.
teins. It can be made double stranded by an independent syn-
The mechanisms by which replication is initiated and con-
thesis event. These different mechanisms will be described in
tinued are different for different viruses.
more detail in the discussions of the different DNA viruses in
DNA polymerases, in general, are unable to initiate a polynu-
Chapter 7.
cleotide chain. They can only extend an existing chain, following
As infection proceeds, most DNA viruses undergo a regu-
instructions from a DNA template. Replication of cellular DNA,
lar developmental cycle, in which transcription of early genes
including that of bacteria, requires the initiation of polynucleotide
is followed by the transcription of late genes. Activation of
chains by a specific RNA polymerase called DNA polymerase
the late genes may result from production of a new RNA
α-primase, or primase for short. The resulting RNA primers are
polymerase or the production of factors that change the activ-
then extended by DNA polymerase. The ribonucleotides in the
ity of existing polymerases so that a new class of promoters
primer are removed after extension of the polynucleotide chain
is recognized. The developmental cycle is, in general, more
as DNA. Removal requires the excision of the ribonucleotides by
elaborate in the larger viruses than in the smaller viruses.
a 5¢ → 3¢ exonuclease, fill-in by DNA polymerase, and sealing
of the nick by ligase. Because DNA polymerases can synthesize
Plus-Strand RNA Viruses
polynucleotide chains only in a 5¢ → 3¢ direction, and cannot ini-
tiate a DNA chain, removal of the RNA primer creates a problem
A simple schematic of the replication of a plus-strand
at the end of a linear chromosome. How is the 5¢end of a DNA
RNA virus is shown in Fig. 1.10. The virus example shown
chain to be generated? The chromosomes of eukaryotic cells have
is enveloped and gives rise to subgenomic RNAs (see later).
special sequences at the ends, called telomeres, that function in
Although the details of RNA replication and virus release are
replication to regenerate ends. The telomeres become shortened
different for other viruses, this scheme is representative of
with continued replication, and eukaryotic cells that lack telomer-
the steps required for gene expression and RNA replication.
ase to repair the telomeres can undergo only a limited number of
Following entry of the genome into the cell, the first event
replication events before they lose the ability to divide.
in replication is the translation of the incoming genomic RNA,
Viruses and bacteria have developed other mechanisms
which is a messenger, to produce proteins required for synthe-
to solve this problem. The chromosomes of bacteria are cir-
sis of antigenomic copies, also called minus strands, of the
cular, so there is no 5¢ end to deal with. Many DNA viruses
genomic RNA. Because the replication cycle begins by trans-
have adopted a similar solution. Many have circular genomes
lating the RNA genome to produce the enzymes for RNA syn-
(e.g., poxviruses, polyomaviruses, papillomaviruses). Others
thesis, the naked RNA is infectious, that is, introduction of the
have linear genomes that cyclize before or during replication
genomic RNA into a susceptible cell will result in a complete
(e.g., herpesviruses). Some DNA viruses manage to replicate
infection cycle. The antigenomic copy of the genome serves
linear genomes, however. Adenoviruses use a virus-encoded
as a template for the production of more plus-strand genomes.
protein as a primer, which remains covalently linked to the
For some plus-strand viruses, the genomic RNA is the only
5¢ end of the linear genome. The single-stranded parvovi-
mRNA produced, as illustrated schematically in Fig. 1.11A.
rus DNA genome replicates via a foldback mechanism in
It is translated into a polyprotein, a long, multifunctional pro-
which the ends of the DNA fold back and are then extended
tein that is cleaved by viral proteases, and sometimes also by
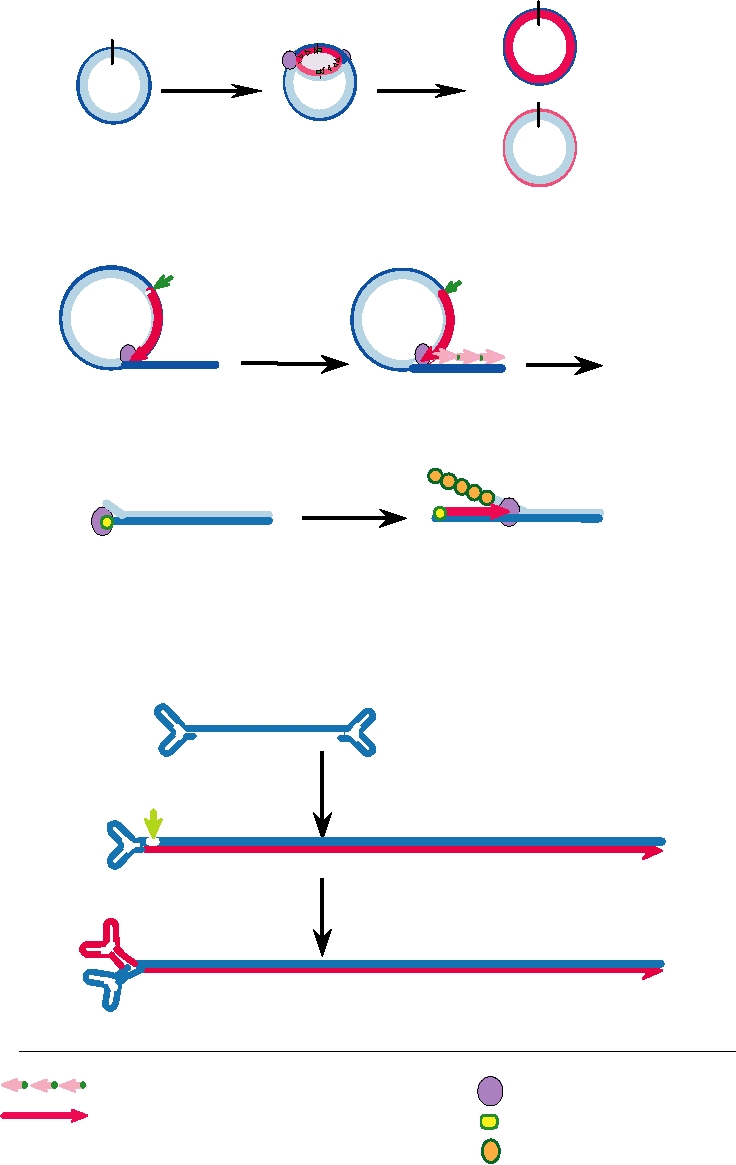
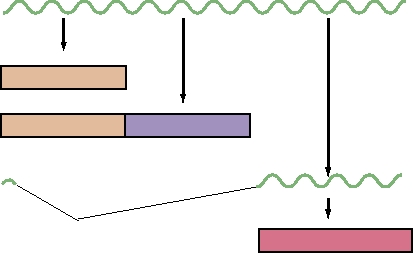
A.
Bidirectional DNA Replication in SV40
Ori
+
Bidirectional
Topoisomerase
DNA synthesis
B.
Rolling Circle DNA Replication in Herpesvirus
3 OH
3 OH
Nick and continuous
Discontinuous DNA
DNA synthesis from 3 OH
synthesis, ligation
Linear DNA
5
5
Concatemer
C.
Adenovirus DNA Replication by Displacement Synthesis
HO
3
3
3
Attachment of preterminal protein
DNA-binding protein coats
Continuous synthesis in 5 to 3 direction
displaced strand
D.
Parvovirus DNA Synthesis by Rolling Hairpin Mechanism
C
c
a
A
Inverted terminal repeats form hairpins
a
A
3
B
b
Elongation from 3 OH
Nick at green arrowhead
C
D
a
d
A
c
b
a
A d
D
a C
B A
B
3
Elongation from nick to left
b
Reform hairpins for primers
c
a
d
A
c
b
a
D
C
A d
D
a C
B A
F
B
Lagging strand with multiple primers
Replication complex
Leading strand with RNA primer
Preterminal protein
DPB, single-strand DNA-
binding protein
IGURE 1.9 Models for DNA replication in various virus groups. Since DNA chains cannot be initiated de novo, viruses
have used a variety of ways to prime new synthesis, such as (A) using RNA primers generated by a primase, (B) elongation
from a 3¢OH formed at a nick in a circular molecule, (C) priming by an attached protein, and (D) priming by hairpins
formed of inverted terminal repeats. Adapted from Flint et al. (2000) Figures 9.8, 9.16, 9.10, and 9.9, respectively.
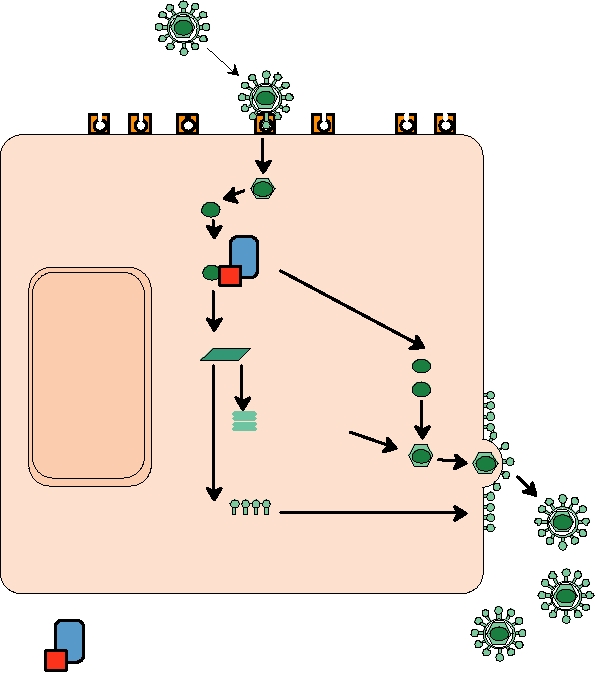
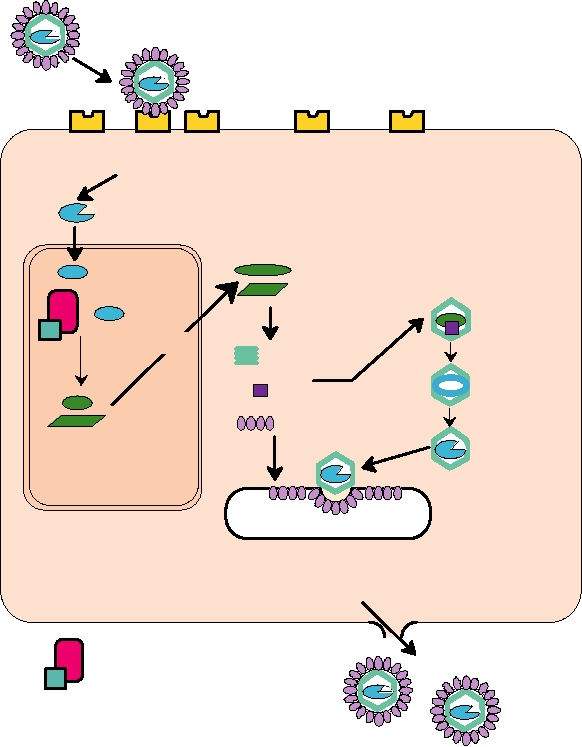
Endocytosis or fusion
Genome RNA
Translation of replicase proteins
Replication
Synthesis of
subgenomic mRNA
mRNA
Translation
Capsid protein
NUCLEUS
Budding
Translation
and modification
Glycoproteins
EUKARYOTIC HOST CELL
Viral RNA synthetase
Host factor(s)
FIGURE 1.10 Replication of an enveloped, plus-strand RNA virus. After the virus attaches to a cellular receptor,
fusion of the virus envelope with the cell plasma membrane or with an endocytic vesicle releases the nucleocapsid into
the cytoplasm. The genome RNA is an mRNA, and is translated on cytoplasmic ribosomes into the proteins required
for RNA synthesis. The synthetase complex can both replicate the RNA to produce new genomes and synthesize viral
subgenomic mRNAs from a minus-strand copy of the genome. The viral structural proteins are then translated from these
subgenomic mRNAs. In the example shown, the capsid protein assembles with the genome RNA to form a capsid, while
the membrane glycoproteins are transported to the cell plasma membrane. In the final maturation step the nucleocapsid
buds out through areas of modified membrane to release the enveloped particle. Adapted from Mims et al. (1993) p. 2.3
and Strauss and Strauss (1997) Figure 2.2.
cellular proteases, to produce the final viral proteins. For other
teins encoded by the virus, one of which is an RNA polymer-
plus-strand RNA viruses, one or more subgenomic mRNAs
"e. Cellular proteins are also components of the synthetase.
are also produced from the antigenomic template (Fig.
All eukaryotic plus-strand RNA viruses replicate in the
1.11B). For these viruses, the genomic RNA is translated into
cytoplasm. There is no known nuclear involvement in their
a polyprotein required for RNA replication (i.e., the synthesis
replication. In fact, where examined, plus-strand viruses will
of the antigenomic template and synthesis of more genomic
even replicate in enucleated cells. However, it is known that
RNA) and for the synthesis of the subgenomic mRNAs. The
for many viruses, virus-encoded proteins are transported to the
subgenomic mRNAs are translated into the structural proteins
nucleus, where they may inhibit nuclear functions. For example,
required for assembly of progeny virions. Some viruses, such
a poliovirus protein cleaves transcription factors in the nucleus.
as the coronaviruses (family Coronaviridae), which produce
multiple subgenomic RNAs, also use subgenomic RNAs to
Minus-Sense and Ambisense RNA Viruses
produce nonstructural proteins that are required for the virus
replication cycle but not for RNA synthesis.
The ambisense RNA viruses and the minus-sense viruses are
The replication of the genome and synthesis of subgenomic
closely related. One family, the Bunyaviridae, even contains
RNAs require recognition of promoters in the viral RNAs by
both types of viruses as members. The ambisense strategy
the viral RNA synthetase. This synthetase contains several pro-
is, in fact, a simple modification of the minus-sense strategy,
A.
Simple Plus-Strand RNA Virus
Template
()
Genome RNA is the
Cap or VPg
(+) only message
Genome
An
(mRNA)
55
3
Viral polyprotein
B.
Complex Plus-Strand RNA Virus
3
5
(+)
Genome
One or more
An
(mRNA)
() subgenomic
Template
RNAs in addition to
genomic RNA
(+)
sg mRNA
An
Viral proteins
C.
Minus-Strand RNA Virus
3
3
(+)
Template
() Subgenomic mRNAs
Genome
synthesized from
sg mRNA
An
An (+) minus strand genome
An
Viral proteins
D.
Ambisense Minus-Strand RNA Virus
Viral proteins
) + (
sg mRNA
An
Subgenomic mRNAs
Template
transcribed from both
Genome
genome and
antigenome RNA
5
sg mRNA
) + (
An
Viral proteins
Synthesis of
Replication
Cap or VPg
Ribosome
subgenomic mRNA
FIGURE 1.11 Schematic of mRNA transcription and translation for the four major types of RNA viruses.
and these viruses are generally lumped together as "negative-
nent of an infectious virion and the naked RNA is not infec-
strand" or "minus-strand" RNA viruses (Table 1.2).
tious if delivered into a cell.
A simple schematic of the replication of a minus-sense
Multiple mRNAs are synthesized by the enzymes present
or ambisense RNA virus is shown in Fig. 1.12. All of these
in the nucleocapsid. Each mRNA is usually monocistronic
viruses are enveloped. After fusion of the virus envelope with
in the sense that it is translated into a single protein, not
a host cell membrane (some enter at the plasma membrane,
into a polyprotein (illustrated schematically in Fig. 1.11C).
some via the endosomal pathway), the virus nucleocapsid
mRNAs are released from the nucleocapsid into the cyto-
enters the cytoplasm. The nucleocapsid is helical (Chapter 2).
plasm, where they are translated. The newly synthesized
It remains intact and the viral RNA is never released from
proteins are required for the replication of the genome.
it. Because the viral genome cannot be translated, the first
Replication of the RNA requires the production of a com-
event after entry of the nucleocapsid must be the synthesis
plementary copy of the genome, as is the case for all RNA
of mRNAs. Thus, the minus-sense or ambisense strategy
viruses, but the antigenomic or vcRNA (for virion comple-
requires that the viral RNA synthetase be an integral compo-
mentary) is distinct from mRNA (Fig. 1.11C). Although
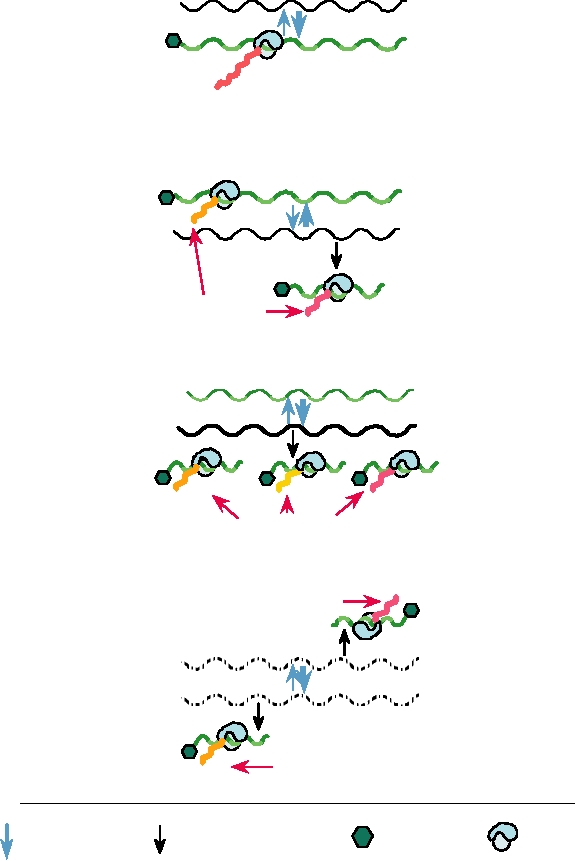
Endocytosis or fusion
mRNA synthesis
mRNAs
Replication
Translation
vcRNA
(+)
Capsid proteins
Replication
Progeny
RNA Genomes
(-)
Translation
Budding
and modification
Glycoproteins
NUCLEUS
EUKARYOTIC HOST CELL
Nucleocapsid with genome RNA(-)
Nucleocapsid with vcRNA (+)
Viral RNA synthetase
FIGURE 1.12 Replication of a typical minus-strand RNA virus. After the virus attaches to a cellular receptor, the
nucleocapsid, containing the viral RNA synthetase, is released into the cytoplasm. The viral synthetase first synthesizes
mRNAs, which are translated into the viral proteins required for synthesis of full-length complementary RNAs
(vcRNAs). These vcRNAs are the templates for minus-strand genome RNA synthesis. Throughout replication, minus-
strand genomes and plus-strand vcRNAs are present in nucleocapsids. Viral mRNAs are also translated into membrane
glycoproteins that are transported to the cell plasma membrane (or in some cases specialized internal membranes). In the
final maturation step, the nucleocapsid buds out through areas of modified membrane to release the enveloped particle.
Adapted from Strauss and Strauss (1997) Figure 2.3 on p. 77.
technically plus sense, it is not translated and is always
The effect is to delay the synthesis of mRNAs that are made
present in nucleocapsids with the associated RNA synthetic
from the antigenomic RNA and thus to introduce a timing
machinery. Replication requires ongoing protein synthesis to
mechanism into the virus life cycle.
supply protein for encapsidation of the nascent antigenomic
The mRNAs synthesized by minus-sense or ambisense
RNA during its synthesis. In the absence of such protein,
viruses differ in several key features from their templates.
the system defaults to the synthesis of mRNAs. The anti-
First, the mRNAs lack the promoters required for encap-
genomic RNA in nucleocapsids can be used as a template to
sidation or replication of the genome or antigenome. Thus,
synthesize genomic RNA if proteins for the encapsidation of
they are not encapsidated and do not serve as templates for
the nascent genomic RNA are available.
the synthesis of minus strand. Second, as befits their func-
In the ambisense viruses, the antigenomic RNA can also
tion as messengers, the mRNAs of most of these viruses are
be used as a template for mRNA (Fig. 1.11D). Thus, ambi-
capped and polyadenylated, whereas genomic and antig-
sense viruses modify the minus-sense strategy by synthe-
enomic RNAs are not. Third, the mRNAs of the viruses in the
sizing mRNA from both the genome and the antigenome.
families Orthomyxoviridae, Arenaviridae, and Bunyaviridae
have 5¢ extensions that are not present in the genome or
Neither the genome nor the antigenome serves as mRNA.
antigenome, which, where well studied, are obtained from cel-
translated by the usual cellular machinery. Thus, the reoviruses
lular mRNAs. Fourth, although most minus-strand and ambi-
share with the minus-strand RNA viruses the attribute that the
sense RNA viruses replicate in the cytoplasm, influenza virus
incoming virus genome remains associated with virus proteins
and bornavirus RNA replication occurs in the nucleus. Thus,
in a core that has the virus enzymatic machinery required to
these RNAs have access to the splicing enzymes of the host.
synthesize RNA, and the first step in replication, following
Two of the mRNAs of influenza viruses are exported in both
entry into a cell, is the synthesis of mRNAs.
an unspliced and a singly spliced version, and bornaviruses
The mRNAs also serve as intermediates in the replication
produce a number of spliced as well as unspliced mRNAs.
of the viral genome and the formation of progeny virions. After
translation, the mRNAs become associated with virus proteins.
At some point, complexes are formed that contain double-
Double-Stranded RNA Viruses
stranded forms of the mRNAs; in these complexes, the 1012
The Reoviridae, the best studied of the double-strand RNA
segments are found in equimolar amounts. These complexes can
viruses, comprise a very large family of viruses that infect ver-
mature into progeny virions. In other words, mRNAs eventually
tebrates, insects, and plants (Table 1.2). The genome consists of
form the plus strands of the double-strand genome segments.
1012 pieces of double-strand RNA. The incoming virus parti-
cle is only partially uncoated. This partial uncoating activates
Retroviruses
an enzymatic activity within the resulting subviral particle or
core that synthesizes an mRNA from each genome fragment.
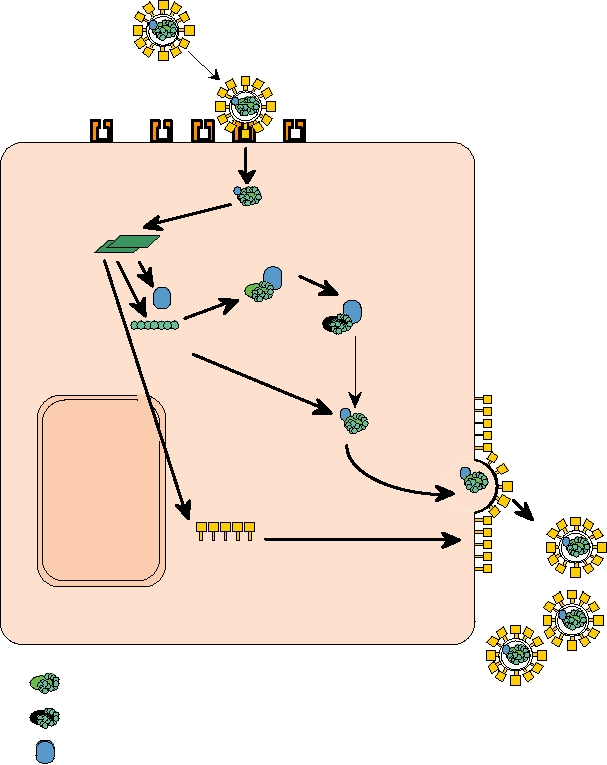
An overview of the replication cycle of a retrovirus is
These mRNAs are extruded from the subviral particle and
shown in Fig. 1.13. The retroviruses are enveloped and enter
Reverse transcription
DNA copy of genome
Genomic
RNAs
Integration
into host DNA
mRNAs
Translation
RNA synthesis
RT
Gag
Budding
Splicing
NUCLEUS
Maturation
Glycoproteins
EUKARYOTIC HOST CELL
FIGURE 1.13 Replication of a retrovirus. After entering the cell the retrovirus RNA genome is reverse transcribed into
double-stranded DNA by RT present in the virion. The DNA copy migrates to the cell nucleus and integrates into the host
genome as the "provirus." Viral mRNAs are transcribed from proviral DNA by host cell enzymes in the nucleus. Both
spliced and unspliced mRNAs are translated into viral proteins in the cytoplasm. The capsid precursor protein, "Gag,"
and RT are translated from full-length RNA. The glycoproteins are translated from spliced mRNA and transported to
the cell plasma membrane. Immature virions containing Gag, RT, and the genome RNA assemble near the modified cell
membrane. The final maturation step involves proteolytic cleavage of Gag by the viral protease and budding to produce
enveloped particles. Adapted from Fields et al. (1996) p. 1786, and Coffin et al. (1997) p. 8.
the cell by fusion, some at the plasma membrane, some at
the viral protease cleaves Gag into several components and
an internal membrane. After entry, the first event is the pro-
also separates the enzymes. Cleavage is essential for the
duction of a double-strand DNA copy of the RNA genome.
assembled virion to be infectious. Thus, current inhibitors of
This requires the activities of the enzymes RT and RNase
HIV target the protease of the virus as well as the RT, both
H, which are present in the virion. RT synthesizes DNA
of which are required for replication, but neither of which is
from either a DNA or an RNA template. RNase H degrades
present in the uninfected cell.
the RNA strand of a DNARNA hybrid and is essential for
The simple retroviruses also produce one spliced mRNA,
reverse transcription of the genome. The mechanism by
which is translated into a precursor for the envelope glycopro-
which the genome is reverse transcribed is complicated and
teins. In some retroviruses, notably the lentiviruses, of which
is described in detail in Chapter 6.
HIV is a member, differential splicing can also lead to the
The double-strand DNA copy of the genome is trans-
production of mRNAs for a number of regulatory proteins.
ported to the nucleus, where it integrates into host DNA.
Integration is essentially random within the host genome and
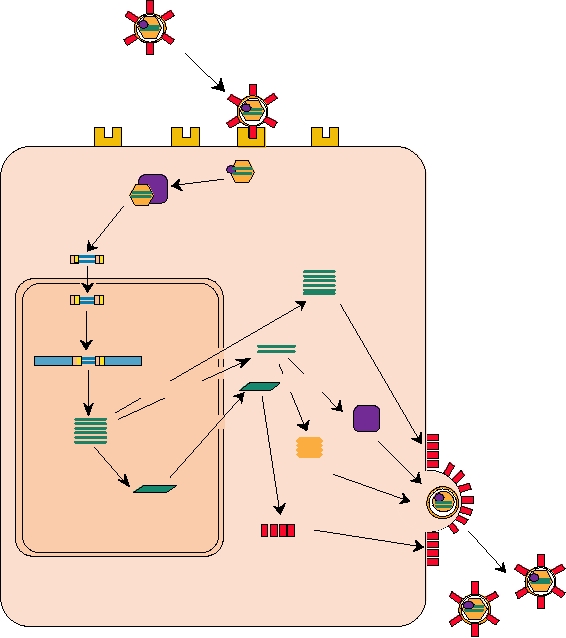
Hepadnaviruses
requires a recombinational event that is catalyzed by another
protein present in the virus, called integrase. The integrated
A schematic of the replication of an hepadnavirus is
DNA copy, called a provirus, is transcribed by cellular RNA
shown in Fig. 1.15. Hepadnaviruses, which are enveloped,
polymerases to produce an RNA that is identical to the viral
have a life cycle that also involves alternation of the infor-
genome. This RNA is exported to the cytoplasm either
mation in the genome between DNA and RNA. The incom-
unspliced or as one or more spliced mRNAs.
ing genome is circular, partially double-stranded DNA. The
The genomic RNA is a messenger for the translation
genome is transported to the nucleus where it is converted to
of a series of polyproteins. These polyproteins contain
a covalently closed, circular, double-strand DNA (cccDNA).
the translation products of genes called gag, pro, and pol.
Unlike the retroviruses, the DNA does not integrate into
Gag (group-specific antigen) proteins form the capsid of
the host genome but persists in the nucleus as a nonrepli-
the virus. Pro is a protease that processes the polyprotein
cating episome. It is transcribed by cellular RNA polymer-
precursors. Pol contains RT, RNase H, and integrase. The
ases, using several different promoters in the cccDNA, to
three genes are immediately adjacent in the genomic RNA,
produce a series of RNAs. These RNAs are exported to the
separated by translation stop codons whose arrangement and
cytoplasm where they serve as mRNAs. One of these RNAs,
number depend on the virus. The arrangement of stop codons
called the pregenomic (pg) RNA, is slightly longer than unit
is described in detail in Chapter 6, and the mechanisms by
length and serves as a template for reverse transcription into
which readthrough of stop codons occurs to produce longer
DNA. Reverse transcription is performed by RT and RNase
polyproteins are described later in this chapter. A simple dia-
H that are translated from the viral mRNAs. It occurs in a
gram of one retrovirus arrangement is shown in Fig. 1.14
core particle assembled from viral capsid proteins, the viral
as an example. In this example, the polyproteins translated
enzymes, and pgRNA. Reverse transcription, described in
from the genomic RNA are Gag and GagProPol. These
detail in Chapter 6, resembles that which occurs in the retro-
two polyproteins assemble with two copies of the virus
viruses, but differs in details. A complete minus-sense DNA
genome to form the capsid of the virus, usually at regions of
is first synthesized by reverse transcription. Second strand
the plasma membrane where virus glycoproteins are present.
plus-sense DNA is then initiated but only partially com-
During and immediately after virus assembly by budding,
pleted, so that the core contains partially double-stranded,
Genome RNA
(gag mRNA)
Translation
Readthrough or
(gag-pol mRNA)
frameshift
translation
gag
pro
Splicing
gag
pol
Env mRNA
Translation
env
FIGURE 1.14 Transcription and translation of the retroviral genome. Three major polyproteins (shown as colored
blocks) are produced. Gag is processed to form the nucleocapsid proteins, Pol contains RT, RNase H, and integrase, and
Env is the precursor to the membrane glycoproteins. The protease, "pro," lies between gag and pol and may be in either
the gag reading frame or the pol reading frame.
Partially ds genome DNA
Repair
RNA
pgRNA
ccc DNA
Encapsidation
mRNA
Translation
+
RT
Transcription
Coat Proteins
+
(-)strandDNA
RT
+
Glycoproteins
Partially
ds DNA
NUCLEUS
Budding into ER
EUKARYOTIC HOST CELL
Release
Host RNA Polymerase
Viral-encoded factor
FIGURE 1.15 Simplified scheme of hepadnavirus replication. In the virion, the partially ds DNA genome consists of
one full-length minus-strand DNA, and a plus-strand DNA of variable length. After the virus enters the cell, the genome
is repaired to a closed circular coiled form (cccDNA) in the nucleus. RNAs, of several sizes, including pregenomic (pg)
RNA of greater than unit length, are transcribed by host RNA polymerases. These mRNAs are exported and translated
into viral proteins in the cytoplasm. The pgRNA is encapsidated, then reverse transcribed into minus-strand DNA. The
last step before budding into the endoplasmic reticulum is the partial synthesis of (+) strand DNA. Scheme derived from
Fields et al. (1996) p. 2709.
circular DNA (i.e., the genome). The core with its DNA can
Cellular Functions Required for Replication
proceed through one of two pathways. Early in infection,
and Expression of the Viral Genome
newly assembled cores may serve to amplify the cccDNA
The relationship between a virus and its host is an intimate
present in the nucleus. These cores contain genomic DNA
one, shaped by a long history of coevolution. Viruses have
and are essentially indistinguishable from cores that enter
small genomes and cannot encode all the functions required
the cytoplasm upon infection by a virion, and their genomic
for successful replication and have borrowed many cellular
DNA can be transported to the nucleus and converted into
proteins as components of their replication machinery. The
cccDNA. Amplification of cccDNA occurs only through an
nature of the interactions between virus proteins and cellular
RNA intermediate, using the pathway just described; there
proteins is an important determinant of the host range and
is no direct replication of the DNA in the nucleus. Later in
pathology of a virus.
infection, the cores mature into virions by budding through
All animal DNA viruses, with the exception of the poxvi-
the endoplasmic reticulum. The switch to budding appears to
ruses, replicate in the nucleus. They make use of the cellular
be driven by the presence of viral envelope proteins.
machinery that exists there for the replication of their DNA
it is clear that such factors can potentially limit the host range
and the transcription of their mRNAs. Some viruses use this
of a virus. The restriction of SV40 in cells that do not make
machinery almost exclusively, whereas others, particularly
a compatible primase was cited earlier in this section. As a
the larger ones, encode their own DNA or RNA polymerases.
second example, the replication of poliovirus in a restricted
However, almost all DNA viruses encode at least a protein
set of cells in the gastrointestinal tract and its profound tro-
required for the recognition of the origin of replication in
pism for neurons if it reaches the CNS was also described
their DNA. The interplay between the viral proteins and the
earlier. These tropisms exhibited by poliovirus do not corre-
cellular proteins can affect the host range of the virus. The
late with the distribution of receptors for the virus, and are a
monkey virus SV40 (family Polyomaviridae) will replicate
result of restrictions on growth after entry of the virus. Thus
in monkey cells but not in mouse cells, whereas the closely
in addition to a requirement for a specific receptor for the
related mouse polyomavirus (also family Polyomaviridae)
virus to enter a cell, there may be a need for specific host
will replicate in mouse cells but not in monkey cells. The
factors to permit replication once a virus enters a cell. The
basis for the host restriction is an incompatibility between
permissivity of a cell for virus replication after its entry, as
the DNA polymerase α-primase of the nonpermissive host
well as the distribution of receptors for a virus, are major
and the T antigen of the restricted virus. T antigens are large
determinants of viral pathogenesis.
multifunctional proteins, one of whose functions is to bind
to the origin of replication. The T antigens of the viruses
Translation and Processing of Viral Proteins
form a preinitiation complex on the viral origin of repli-
cation, which then recruits the primase into the complex.
Viral mRNAs are translated by the cellular translation
Because the preinitiation complex containing SV40 T anti-
machinery. Most mRNAs of animal viruses are capped and
gen cannot recruit the mouse primase to form an initiation
polyadenylated. Thus, the translation pathways are the same
complex, SV40 DNA replication does not occur in mouse
as those that operate with cellular mRNAs, although many
cells. However, replication will occur in mouse cells if they
viruses interfere with the translation of host mRNAs to give
are transfected with the gene for monkey primase. Similarly,
the viral mRNAs free access to the translation machinery.
monkey primase is not recruited into the complex contain-
However, there are mechanisms of translation and process-
ing mouse polyoma virus T antigen and mouse polyoma
ing used by some viruses that have no known cellular coun-
virus does not replicate in monkey cells.
terpart. These appear to have evolved because of the special
In the case of RNA viruses, there is no preexisting cel-
problems faced by viruses and are described below.
lular machinery to replicate their RNA, and all RNA viruses
must encode at least an RNA-dependent RNA polymerase.
Cap-Independent Translation of Viral mRNAs
This RNA polymerase associates with other viral and host
A 5¢-terminal cap on an mRNA is normally required for
proteins to form an RNA replicase complex, which has the
ability to recognize promoters in the viral RNA as starting
its translation. A cellular cap-binding protein binds the cap as
points for RNA synthesis. Early studies on the RNA repli-
part of the translation initiation pathway. The mRNA is then
case of RNA phage Qβ showed that three cellular proteins
scanned by the initiation complex, starting at the cap, and
were associated with the viral RNA polymerase and were
translation begins at a downstream AUG start codon that is
required for the replication of Qβ RNA. These three pro-
present in a favorable context. However, some viruses, such
teins, ribosomal protein S1 and two translation elongation
as poliovirus and other members of the Picornaviridae and
factors EF-Ts and EF-Tu, all function in protein synthesis in
hepatitis C virus (genus Hepacivirus, family Flaviviridae),
the cell. The virus appropriates these three proteins in order
have uncapped mRNAs and use another mechanism for
the initiation of translation. The 5¢ nontranslated regions
to assemble an active replicase complex. Recent studies on
animal and plant RNA viruses have shown that a variety of
(NTRs) of the RNAs of two picornaviruses, poliovirus and
cellular proteins also appear to be required for their tran-
encephalomyelitis virus (EMCV), are illustrated schemati-
cally in Fig. 1.16. These 5¢ NTRs are long--more than 700
scription and replication. One interesting finding is that the
nucleotides. Within this 5¢ region is a sequence of about 400
animal equivalents of EF-Ts and EF-Tu are required for the
activity of the replicase of vesicular stomatitis virus. This
nucleotides called an IRES (internal ribosome entry site).
suggests that the association of these two translation factors
Ribosomes bind to the IRES and initiate translation in a cap-
with viral RNA replicases is ancient. Several other cellular
independent fashion. It is known that the secondary structure
proteins have also been found to be associated with viral
of the IRES is critical for its function and that the position
RNA polymerases or with viral RNA during replication, but
of the initiating AUG with respect to the IRES is important.
evidence for their functional role is incomplete.
Interestingly, IRES elements may have entirely different
Although our knowledge of the nature of host factors
sequences in different viruses and, hence, different apparent
involved in the replication of viral genomes and the interplay
higher order structures. Comparison of the IRES elements of
between virus-encoded and host cell proteins is incomplete,
poliovirus and EMCV in Fig. 1.16 illustrates the differences
I
560
EMCV (Type II) IRES
nitiating
A
AUG codon
620
nt 834
D
30
70
300
M
F
C
L
360
460
100
N
800
I3
260 2
120
B
K
E
G
700
420
H
J
VPg
PV (Type I) IRES
Poly (C)
Pyrimidine-rich tract
IV
388
Spacer (15-20 nts)
V
87
AUG
AUG codon
IRES
VI
II
I
III
Initiating
462
AUG codon
nt 743
547
5
110
620
AUG 3
5
162
220
123 nt
FIGURE 1.16 Schematic diagram of the secondary structures in the 5¢-nontranslated regions of encephalomyelocarditis
virus (EMCV) and poliovirus (PV). The internal ribosome entry sites, or IRES elements, are shaded. There is an element
at the 3¢ border of the IRES consisting of a pyrimidine-rich tract (box with diagonal hatching), a spacer (magenta line)
(usually 1520 nucleotides) and an AUG codon. In the Type II IRES this AUG is the initiating codon, but in a Type I IRES
a second codon, in this case 123 nucleotides further down, is used for initiation. Adapted from Wimmer et al. (1993)
p. 374 with permission.
in apparent structure. Yet they promote internal initiation in
of a 3¢ gene. Since it is possible to construct polycistronic
a similar fashion, and the IRES elements of poliovirus and
mRNAs using IRES elements, it is somewhat puzzling that
EMCV can be exchanged to yield a viable virus.
animal viruses have never used them to do so.
The IRES elements of viruses are always found within
Using an IRES allows a virus to preferentially translate
the 5¢ NTR. However, if an IRES is placed in the middle of
viral mRNAs. Thus, for example, poliovirus blocks cap-
an mRNA, it will function to initiate translation. True poly-
dependent translation after infection by cleaving the cellular
cistronic mRNAs have been constructed by placing multiple
cap-binding protein. By blocking cap-dependent translation,
IRES elements into mRNAs, or by combining cap-dependent
most host mRNAs cannot be translated (although IRESs
translation of a 5¢ gene with an IRES-dependent translation
are known to exist in some eukaryotic mRNAs), whereas
viral mRNAs are not affected. This reserves the translation
shows the -1 frameshifting that occurs between the gag
machinery for translation of viral mRNAs and also blocks
and pol genes of avian leukosis virus (ALV). Frameshifting
many host defense mechanisms (Chapter 10).
occurs at a precise sequence known as a "slippery" sequence,
shown in green. Slippery sequences have short strings of the
same nucleotide and are often rich in A and U. For ALV,
Ribosomal Frameshifting
termination at the UAG indicated with the asterisk produces
Retroviruses, many plus-strand RNA viruses, and certain
a polyprotein that contains the sequences for Gag and Pro.
other viruses expand the range of polyproteins produced by
Frameshifting results in movement into the -1 frame at this
having long open reading frames (ORFs) interrupted by stop
point so that the codon following UUA (leucine) becomes
codons. Termination of the polyprotein at the stop codon
AUA (isoleucine) rather than UAG (stop), and transla-
leads to production of a truncated polyprotein having cer-
tion of the downstream gene (pol) follows to produce the
tain functions, whereas readthrough leads to the production
polyprotein GagProPol.
of a longer polyprotein with additional functions. This was
Frameshifting usually requires a structural feature, such
illustrated in Fig. 1.14 for a retrovirus. Two mechanisms
as a hairpin, downstream of the slippery sequence in order
are used by different viruses to ignore the stop codon. The
to slow the ribosome at this point and allow more time for
first is simply to read the stop codon as sense. In this case
the frameshifting to take place. The structure that is required
the downstream sequences are in the same reading frame as
for frameshifting in a yeast RNA virus called L-A is shown
the upstream sequences. The mechanism of readthrough is
in Fig. 1.18. The sizable hairpin just downstream of the slip-
thought to involve wobble in the third codon position that
pery sequence is further stabilized by a pseudoknot. In this
allows a tRNA to bind to and insert an amino acid at the
case the slippery sequence, in which frameshifting takes
stop codon position. Both amber (UAG) and opal (UGA)
place, occurs about 110 nucleotides upstream of the stop
codons have been found in different readthrough posi-
codon for the upstream reading frame (Gag protein reading
tions. Readthrough efficiency is variable, although usually
frame). Thus, these 110 nucleotides are translated in two dif-
between 5 and 20%.
ferent reading frames: the Gag reading frame if frameshift-
The second mechansim for ignoring the stop codon
ing does not occur, or the Pol reading frame if frameshifting
is ribosomal frameshifting in which the reading frame is
does occur. The sequence illustrated in Fig. 1.18 is the mini-
shifted into the plus 1 or minus 1 frame upstream of the stop
mum sequence required for frameshifting. It can be placed
codon. In this case, the downstream sequences are in a dif-
in the middle of an unrelated mRNA and 1 frameshifting
ferent reading frame from the upstream sequences. Fig. 1.17
will occur.
Ribosomal Frameshift in Avian Leukosis Virus
3
5
Protease
N L3 *
L
T
RNA sequence
UUGACAAAUUUAUAGGGAGGGCC
Polymerase
I
G
R
A
Pro-Pol fusion protein
L
T
N
L I
G
R
A
Mechanism of the (-1) Frameshift
I
3
Ribosome
5
UAU
Peptide
N
L
I
Peptide
N
L
Stop
U
AAU
UAU
5
AAU
XA AAU
UUA UAG GGXXXX
X
AAA
UUU
AUA GGG XXXX
t-RNA slips on message as
mRNA shifts one position to
the right
FIGURE 1.17
Proposed mechanism of the (-1) ribosomal frameshift that occurs in ALV (avian leukosis virus). The
slippery sequence is shown in green. The asterisk identifies the UAG codon that terminates the upstream (GagPro)
ORF. Frameshifting is thought to require a pseudoknot downstream of the "slippery sequence" (illustrated in Fig. 1.18).
Adapted from Fields et al. (1996) p. 577 and Goff (1997) Figure 3.15 on p. 156.
teases, of which chymotrypsin is a well-studied example,
. nt 2025
Stem 2
G
3
G
..
have a catalytic triad composed of histidine, aspartic acid,
G U C GC
U
CC
G
A C
and serine, which form the active site of the enzyme. Where
Pseudoknot
U
G
structures have been determined, the viral proteases possess a
C G
A
nt 1983
fold related to that of chymotrypsin and possess an active site
A
G
U
C
C
G
with geometry identical to that of chymotrypsin. The structure
A
U
A
of protease 3Cpro of a rhinovirus (family Picornaviridae) is
U
A
U
U
A
shown in Fig. 1.19A as an example (another example is shown
C
C
U
Stem 1 U
G
C
in Chapter 2, Fig. 2.14B). Although most viral serine-type
G
C
A
G
C
proteases have a catalytic triad composed of histidine, aspar-
A
A
U
U
A
G
tic acid, and serine, the rhinovirus protease has an active site
nt 1951
G
C
A
Slippery site
5
G
C
composed of histidine, glutamic acid, which replaces aspartic
G G
U
...CUCAGCAGGGUUUAGGAG
nt 2005
acid, and cysteine, which replaces serine. The replacement of
Gag ORF
the active site serine by cysteine causes problems with nomen-
Pol ORF
clature. Here we use serine protease (or serine-type protease)
FIGURE 1.18 The site in L-A RNA that promotes 1 ribosomal
to refer to a protease whose structure and active site geometry
frameshifting to form the GagPol fusion protein. The pseudoknot makes
are related to those of the animal serine proteases, rather than
the ribosome pause over the slippery site (nt 19581964 shown in green).
as a description of the catalytic amino acid. These similari-
The gag and pol ORFs overlap for 130 nucleotides and in the absence of
ties in structure make it likely that the viruses acquired the
frameshifting the Gag protein terminates at nt 2072. Adapted from Fields
et al. (1996) p. 566.
protease from a host and modeled it to fit their own needs.
A second group of proteases is related to the papain-like
The efficiency of frameshifting is variable and depends
enzymes of plants and animals. These proteases have a cata-
on the frameshifting sequences. Thus, the efficiency of
lytic dyad consisting of histidine and cysteine. A third residue
frameshifting can be modulated by mutations in the virus.
is sometimes considered a part of the active site (as described
For example, the frameshifting sequence in Fig. 1.18 results
later) and the active site is then referred to as a catalytic triad.
in frameshifting about 2% of the time. Changing the slip-
Model folding studies have suggested that these viral papain-
pery sequence from GGGUUU to UUUUUU results in
like proteases are related to the cellular counterparts. Such
frameshifting 12% of the time. Frameshifting efficiencies of
proteases are found in many plus-strand RNA viruses and
from 2% to more than 20% have been observed at different
in adenoviruses. The structure of the adenovirus papain-like
frameshift sites in different viruses.
protease is shown in Fig. 1.19B. Notice that the structure of this
enzyme is very different from the serine-type protease of rhino-
viruses in Fig. 1.19A, even though the active site is composed
Processing of Viral Polyproteins
of the same three residues, Cys, His, and Glu. In addition, the
Many viruses produce polyproteins, as described earlier
sequence of the active site residues in the linear amino acid
in this chapter. These polyproteins are cleaved into individ-
sequence of serine-type proteases is His, Glu, Cys, whereas the
ual proteins by viral enzymes, with the exception of precur-
order is Cys, His, Glu in the adenovirus protease and in papain
sors to viral envelope proteins, which have access to cellular
(where the active site contains Asn rather than Glu).
enzymes present in subcellular compartments. The use of
The retroviruses encode a protease to process the Gag
viral enzymes to process polyproteins may be due in part to a
and GagPol polyproteins during virus maturation. The pro-
lack of appropriate proteases in the cytoplasm of eukaryotes.
tease is related to the aspartate proteases of animals, which
However, the use of a viral protease also allows the virus to
include pepsin, renin, and cathepsin D. The structure of
fine-tune the processing events, and this control is often used
the HIV protease is shown in Fig. 1.19C. The active site of
to regulate the viral replication cycle.
aspartate proteases consists of two aspartate residues. In the
The viral proteases are components of the translated poly-
animal enzymes, the two aspartate residues are present in
proteins. Some cleavages catalyzed by these proteases can
a single polypeptide chain that folds to bring the aspartate
occur in cis, in a monomolecular reaction in which the poly-
residues together to form the active site. In HIV, the active
protein cleaves itself. Other cleavages occur in trans, whereby
site is formed by dimerization of two Pro protein monomers
a polyprotein, or a cleaved product containing the protease,
of about 100 residues, each of which contributes one of the
cleaves another polyprotein in a bimolecular reaction. In many
aspartate residues to the active site.
cases the series of cleavages effected by a protease proceeds in
a defined manner and serves to regulate the virus life cycle.
Assembly of Progeny Virions
Three types of virus proteases have been found in different
viruses. Proteases related to the serine proteases of animals
The last stage in the virus life cycle is the assembly of
are common in animal RNA viruses. The animal serine pro-
progeny virions and their release from the infected cell. The
Search WWH :